A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
Tizon, L., Otero, J.M., Prazeres, V.F., Llamas-Saiz, A.L., Fox, G.C., van Raaij, M.J., Lamb, H., Hawkins, A.R., Ainsa, J.A., Castedo, L., Gonzalez-Bello, C.(2011) J Med Chem 54: 6063-6084
- PubMed: 21780742
- DOI: https://doi.org/10.1021/jm2006063
- Primary Citation of Related Structures:
2Y71, 2Y76, 2Y77 - PubMed Abstract:
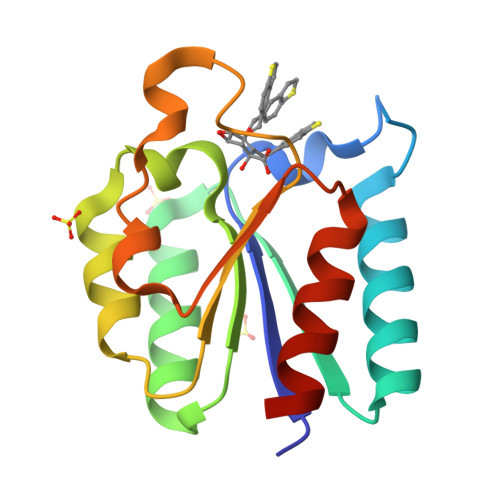
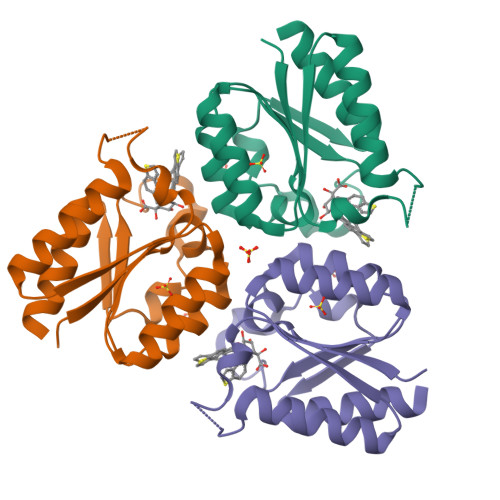
The synthesis of high-affinity reversible competitive inhibitors of Mycobacterium tuberculosis type II dehydroquinase, an essential enzyme in Mycobacterium tuberculosis bacteria, is reported. The inhibitors reported here are mimics of the enol intermediate and the effect of substitution on C2 was studied. The crystal structures of Mycobacterium tuberculosis type II dehydroquinase in complex with three of the reported inhibitors are also described. The results show that an aromatic substituent on C2 prevents the closure of the active site by impeding the hydrogen-bonding interaction of Arg108 with the essential Tyr24 of the flexible loop, the residue that initiates catalysis. Chemical modifications of the reported acids were also carried out to improve internalization into Mycobacterium tuberculosis through an ester prodrug approach. Propyl esters proved to be the most efficient in achieving optimal in vitro activities.
Organizational Affiliation:
Centro Singular de Investigación en Química Biológica y Materiales Moleculares, Universidad de Santiago de Compostela, calle Jenaro de la Fuente s/n, 15782 Santiago de Compostela, Spain.