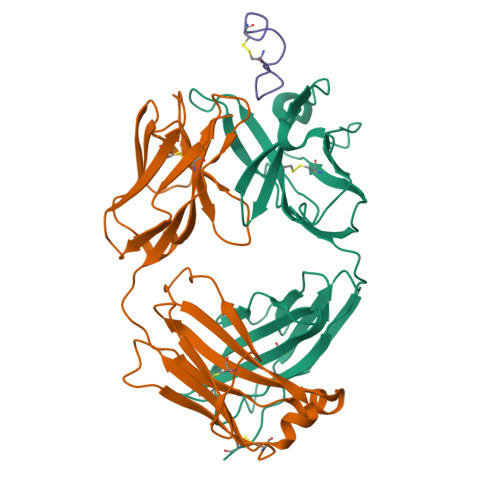
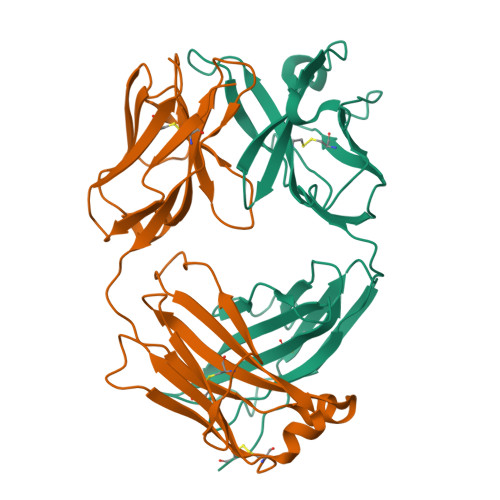
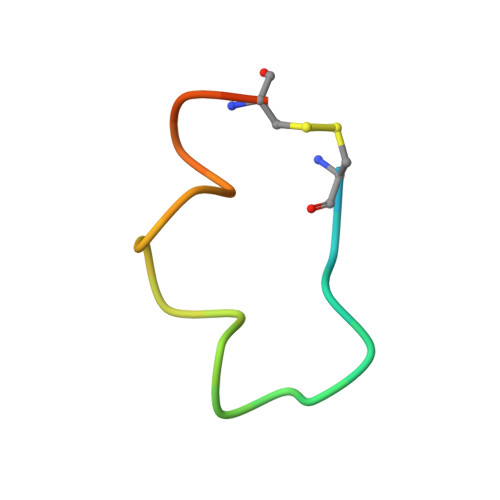
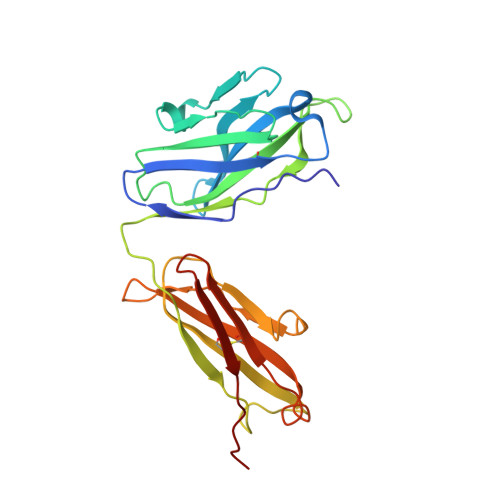
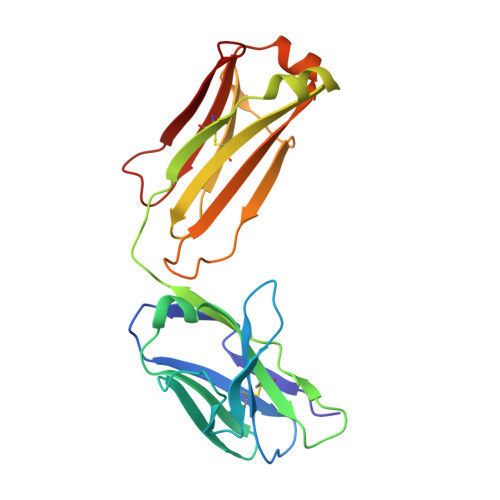
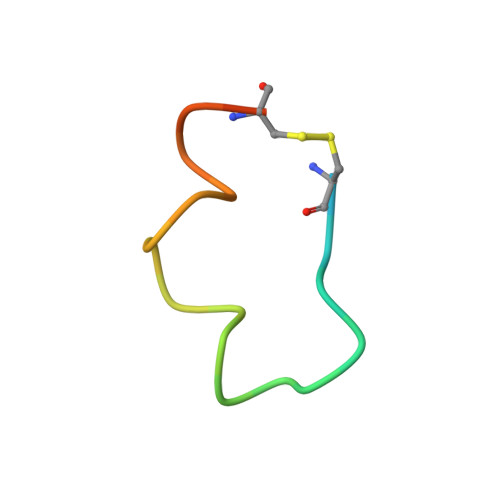
Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Du, J., Wang, H., Zhong, C., Peng, B., Zhang, M., Li, B., Hou, S., Guo, Y., Ding, J.(2008) Mol Immunol 45: 2861-2868
- PubMed: 18346788
- DOI: https://doi.org/10.1016/j.molimm.2008.01.034
- Primary Citation of Related Structures:
3BKY - PubMed Abstract:
Anti-CD20 monoclonal antibodies have been proven to be efficient in the treatment of certain B-cell lymphomas and autoimmune diseases. Intriguingly, these antibodies seem to exert diverse functions with narrow epitope specificity. This study is to investigate the molecular basis of the fine specificity of 2H7 derived antibodies which are of great therapeutic potential. We show that chimeric 2H7 (C2H7) can mediate complement dependent cytotoxicity and antibody-dependent cellular cytotoxicity effects on CD20 positive human Burkitt lymphoma cells and the Fab fragment can well recognize and bind to an epitope peptide of the extracellular loop of CD20. The crystal structure of C2H7 in complex with the CD20 epitope peptide was determined at 2.6A resolution. The bound peptide displays a circular conformation and the binding specificity is mainly contributed by the (170)ANPS(173) motif and the disulfide bond of the peptide which maintains the unique conformation of the peptide. Compared with the complex structure of another anti-CD20 monoclonal antibody Rituximab with the same epitope peptide which was previously determined, the major differences lie in the CDR loop H3 of C2H7 which stretches outward against the interface. Correspondingly, the pocket which accommodates the peptide becomes wider and the peptide moves toward loop H3 and thus is more distant from loops H1 and H2. The hydrogen-bonding interactions are also quite different from those observed in the Rituximab-epitope peptide complex, and both the hydrophilic and hydrophobic interactions are less intense. Our data not only reveal the molecular basis for the fine specificity of C2H7 to CD20, but also provide valuable information for further improvement of antibodies derived from 2H7.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China.