2-amino-1,3-thiazol-4(5H)-ones as potent and selective 11beta-hydroxysteroid dehydrogenase type 1 inhibitors: enzyme-ligand co-crystal structure and demonstration of pharmacodynamic effects in C57Bl/6 mice.
Johansson, L., Fotsch, C., Bartberger, M.D., Castro, V.M., Chen, M., Emery, M., Gustafsson, S., Hale, C., Hickman, D., Homan, E., Jordan, S.R., Komorowski, R., Li, A., McRae, K., Moniz, G., Matsumoto, G., Orihuela, C., Palm, G., Veniant, M., Wang, M., Williams, M., Zhang, J.(2008) J Med Chem 51: 2933-2943
- PubMed: 18419108
- DOI: https://doi.org/10.1021/jm701551j
- Primary Citation of Related Structures:
3BYZ - PubMed Abstract:
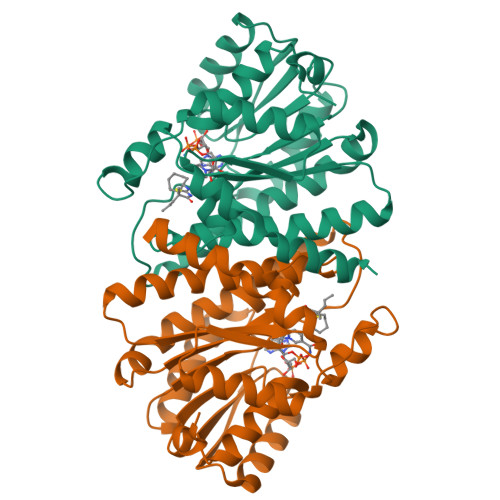
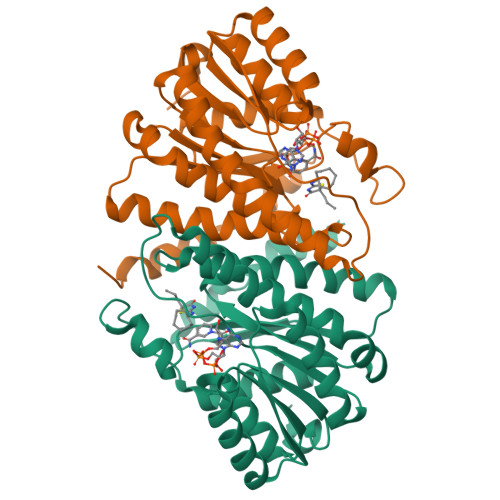
11beta-hydroxysteroid dehydrogenase type 1 (11beta-HSD1) has attracted considerable attention during the past few years as a potential target for the treatment of diseases associated with metabolic syndrome. In our ongoing work on 11beta-HSD1 inhibitors, a series of new 2-amino-1,3-thiazol-4(5 H)-ones were explored. By inserting various cycloalkylamines at the 2-position and alkyl groups or spirocycloalkyl groups at the 5-position of the thiazolone, several potent 11beta-HSD1 inhibitors were identified. An X-ray cocrystal structure of human 11beta-HSD1 with compound 6d (Ki=28 nM) revealed a large lipophilic pocket accessible by substitution off the 2-position of the thiazolone. To increase potency, analogues were prepared with larger lipophilic groups at this position. One of these compounds, the 3-noradamantyl analogue 8b, was a potent inhibitor of human 11beta-HSD1 (Ki=3 nM) and also inhibited 11beta-HSD1 activity in lean C57Bl/6 mice when evaluated in an ex vivo adipose and liver cortisone to cortisol conversion assay.
Organizational Affiliation:
Biovitrum AB, Stockholm, Sweden. lars.johansson@biovitrum.com