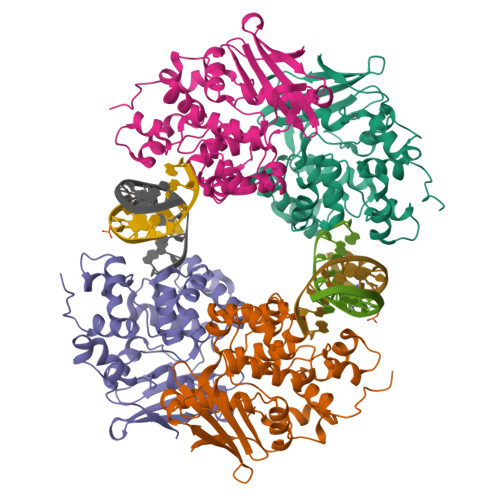
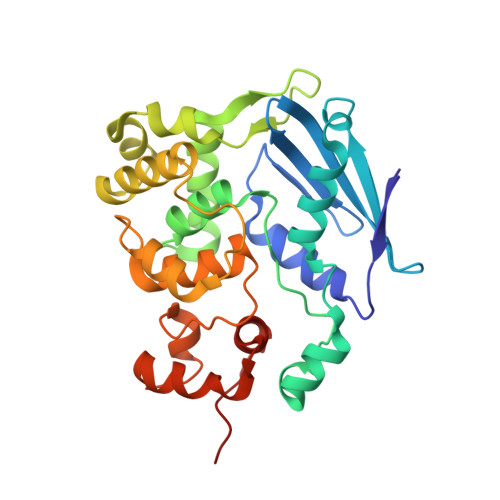
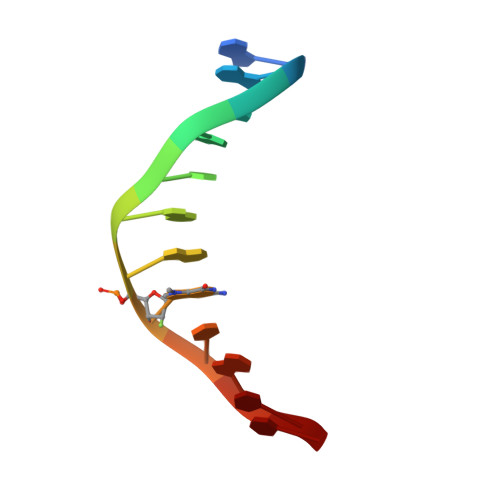
Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
Lee, S., Bowman, B.R., Ueno, Y., Wang, S., Verdine, G.L.(2008) J Am Chem Soc 130: 11570-11571
- PubMed: 18686953
- DOI: https://doi.org/10.1021/ja8025328
- Primary Citation of Related Structures:
3D4V - PubMed Abstract:
The predominant product of aberrant DNA methylation is the genotoxic lesion N7-methyl-2'-deoxyguanosine (m7dG). M7dG is recognized and excised by lesion-specific DNA glycosylases, namely AlkA in E. coli and Aag in humans. Structural studies of m7dG recognition and catalysis by these enzymes have been hampered due to a lack of efficient means by which to incorporate the chemically labile m7dG moiety site-specifically into DNA on a preparative scale. Here we report a solution to this problem. We stabilized the lesion toward acid-catalyzed and glycosylase-catalyzed depurination by 2'-fluorination and toward base-catalyzed degradation using mild, nonaqueous conditions in the DNA deprotection reaction. Duplex DNA containing 2'-fluoro-m7dG (Fm7dG) cocrystallized with AlkA as a host-guest complex in which the lesion-containing segment of DNA was nearly devoid of protein contacts, thus enabling the first direct visualization of the N7-methylguanine lesion nucleobase in DNA. The structure reveals that the base-pairing mode of Fm7dG:C is nearly identical to that of G:C, and Fm7dG does not induce any apparent structural disturbance of the duplex structure. These observations suggest that AlkA and Aag must perform a structurally invasive interrogation of DNA in order to detect the presence of intrahelical m7dG lesions.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, USA.