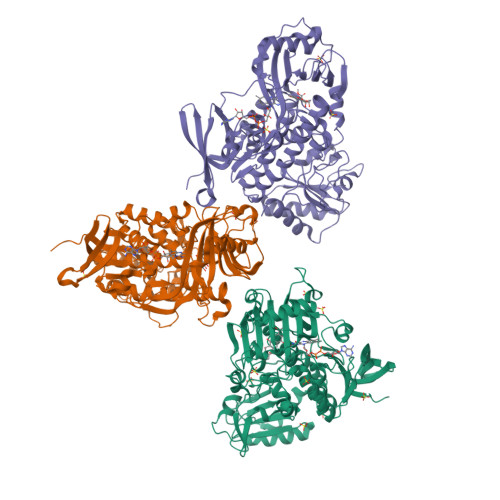
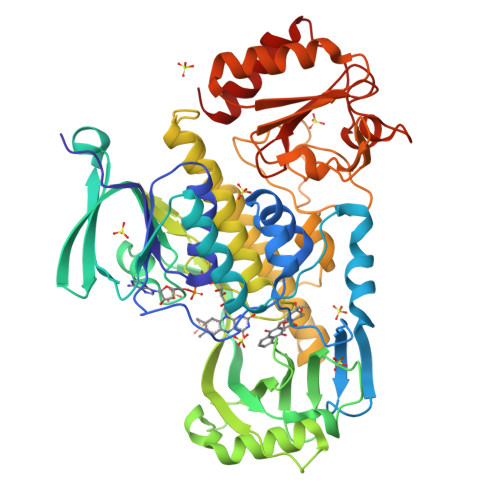
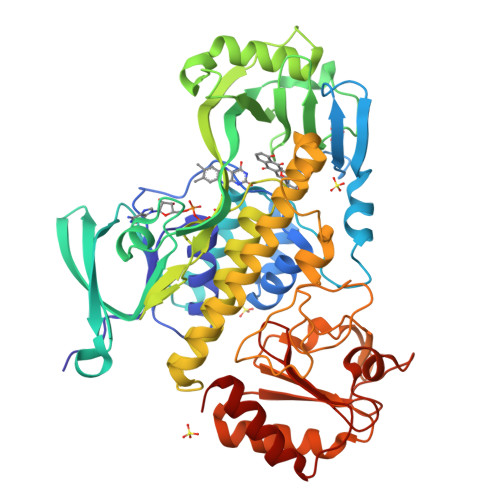
Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
Lindqvist, Y., Koskiniemi, H., Jansson, A., Sandalova, T., Schnell, R., Liu, Z., Mantsala, P., Niemi, J., Schneider, G.(2009) J Mol Biology 393: 966-977
- PubMed: 19744497
- DOI: https://doi.org/10.1016/j.jmb.2009.09.003
- Primary Citation of Related Structures:
3IHG - PubMed Abstract:
In the biosynthesis of several anthracyclines, aromatic polyketides produced by many Streptomyces species, the aglycone core is modified by a specific flavin adenine dinucleotide (FAD)- and NAD(P)H-dependent aklavinone-11-hydroxylase. Here, we report the crystal structure of a ternary complex of this enzyme from Streptomyces purpurascens, RdmE, with FAD and the substrate aklavinone. The enzyme is built up of three domains, a FAD-binding domain, a domain involved in substrate binding, and a C-terminal thioredoxin-like domain of unknown function. RdmE exhibits structural similarity to aromatic hydroxylases from the p-hydroxybenzoate hydroxylase family, but unlike most other related enzymes, RdmE is a monomer. The substrate is bound in a hydrophobic pocket in the interior of the enzyme, and access to this pocket is provided through a different route than for the isoalloxazine ring of FAD-the backside of the ligand binding cleft. The architecture of the substrate binding pocket and the observed enzyme-aklavinone interactions provide a structural explanation for the specificity of the enzyme for non-glycosylated substrates with C9-R stereochemistry. The isoalloxazine ring of the flavin cofactor is bound in the "out" conformation but can be modeled in the "in" conformation without invoking large conformational changes of the enzyme. This model places the flavin ring in a position suitable for catalysis, almost perpendicular to the tetracyclic ring system of the substrate and with a distance of the C4a carbon atom of the isoalloxazine ring to the C-11 carbon atom of the substrate of 4.8 A. The structure suggested that a Tyr224-Arg373 pair might be involved in proton abstraction at the C-6 hydroxyl group, thereby increasing the nucleophilicity of the aromatic ring system and facilitating electrophilic attack by the perhydroxy-flavin intermediate. Replacement of Tyr224 by phenylalanine results in inactive enzyme, whereas mutants at position Arg373 retain catalytic activity close to wild-type level. These data establish an essential role of residue Tyr224 in catalysis, possibly in aligning the substrate in a position suitable for catalysis.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm S-171 77, Sweden.