Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Kallio, J.P., Gasparetti, C., Andberg, M., Boer, H., Koivula, A., Kruus, K., Rouvinen, J., Hakulinen, N.(2011) FEBS J 278: 2283-2295
- PubMed: 21535408
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08146.x
- Primary Citation of Related Structures:
3PPS - PubMed Abstract:
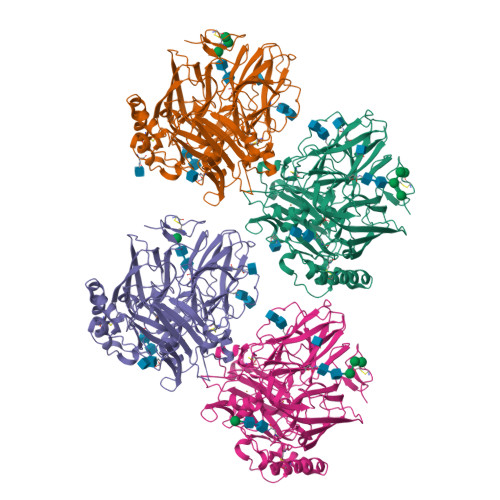
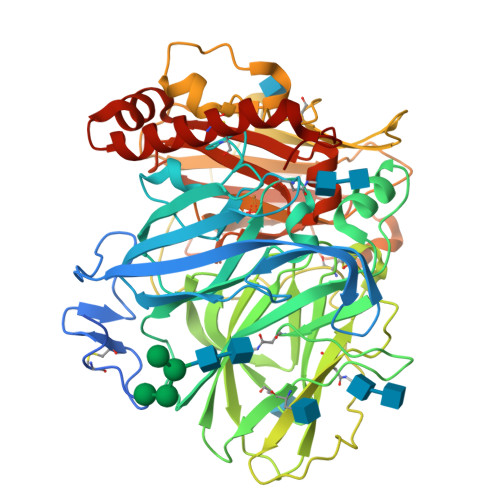
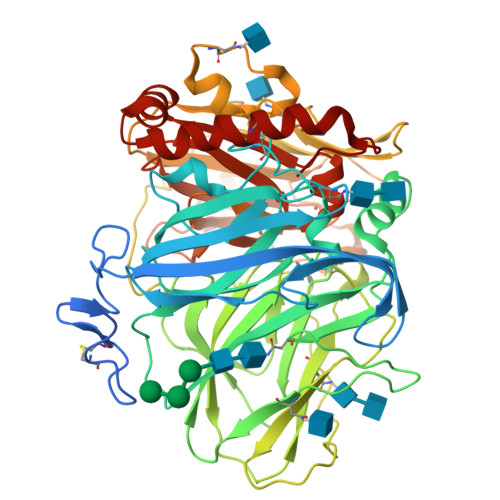
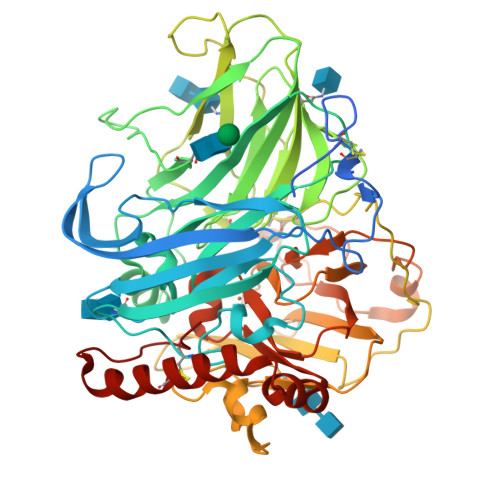
Laccases are copper-containing enzymes used in various applications, such as textile bleaching. Several crystal structures of laccases from fungi and bacteria are available, but ascomycete types of fungal laccases (asco-laccases) have been rather unexplored, and to date only the crystal structure of Melanocarpus albomyces laccase (MaL) has been published. We have now solved the crystal structure of another asco-laccase, from Thielavia arenaria (TaLcc1), at 2.5 Å resolution. The loops near the T1 copper, forming the substrate-binding pockets of the two asco-laccases, differ to some extent, and include the amino acid thought to be responsible for catalytic proton transfer, which is Asp in TaLcc1, and Glu in MaL. In addition, the crystal structure of TaLcc1 does not have a chloride attached to the T2 copper, as observed in the crystal structure of MaL. The unique feature of TaLcc1 and MaL as compared with other laccases structures is that, in both structures, the processed C-terminus blocks the T3 solvent channel leading towards the trinuclear centre, suggesting a common functional role for this conserved 'C-terminal plug'. We propose that the asco-laccases utilize the C-terminal carboxylic group in proton transfer processes, as has been suggested for Glu498 in the CotA laccase from Bacillus subtilis. The crystal structure of TaLcc1 also shows the formation of a similar weak homodimer, as observed for MaL, that may determine the properties of these asco-laccases at high protein concentrations.
Organizational Affiliation:
Department of Chemistry, University of Eastern Finland, Joensuu, Finland.