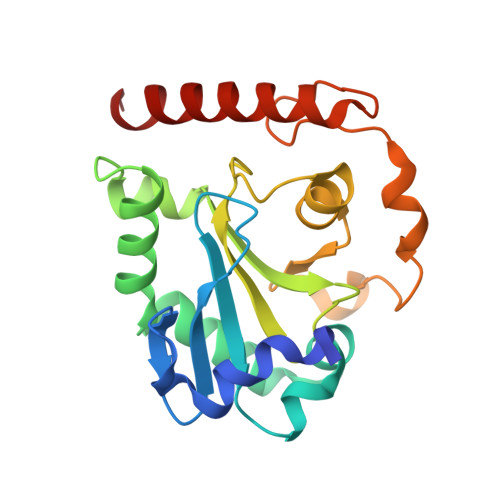
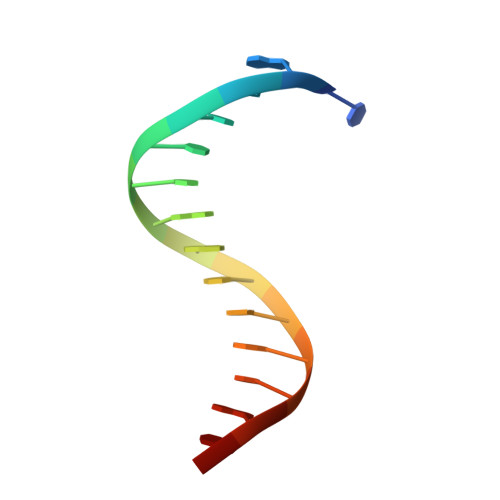
Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Gong, Y., Zhu, D., Ding, J., Dou, C., Ren, X., Gu, L., Jiang, T., Wang, D.(2011) Nat Struct Mol Biol 18: 1297-1299
- PubMed: 21984208
- DOI: https://doi.org/10.1038/nsmb.2145
- Primary Citation of Related Structures:
3SP4, 3SPD, 3SPL - PubMed Abstract:
Aprataxin is a DNA deadenylase that resolves DNA 5'-AMP termini and reverses abortive DNA ligation. The crystal structures of Schizosaccharomyces pombe aprataxin Hnt3 in its apo form and in complex to dsDNA and dsDNA-AMP reveal how Hnt3 recognizes and processes 5'-adenylated DNA in a structure-specific manner. The bound DNA adopts a 5'-flap conformation that facilitates 5'-AMP access to the active site, where AMP cleavage occurs by a canonical catalytic mechanism.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.