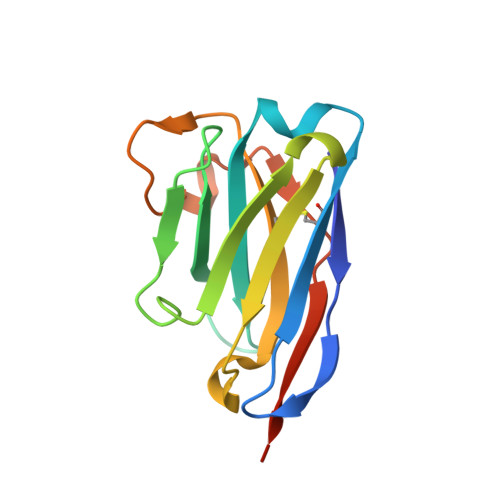
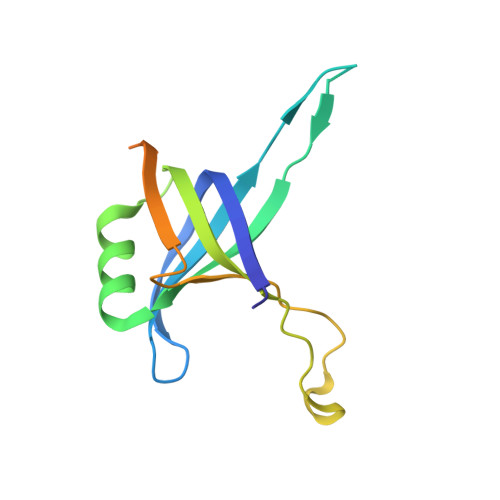
Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody.
Park, Y.J., Pardon, E., Wu, M., Steyaert, J., Hol, W.G.(2012) Nucleic Acids Res 40: 1828-1840
- PubMed: 22039098
- DOI: https://doi.org/10.1093/nar/gkr867
- Primary Citation of Related Structures:
3STB - PubMed Abstract:
The parasite Trypanosoma brucei, the causative agent of sleeping sickness across sub-Saharan Africa, depends on a remarkable U-insertion/deletion RNA editing process in its mitochondrion. A approximately 20 S multi-protein complex, called the editosome, is an essential machinery for editing pre-mRNA molecules encoding the majority of mitochondrial proteins. Editosomes contain a common core of twelve proteins where six OB-fold interaction proteins, called A1-A6, play a crucial role. Here, we report the structure of two single-strand nucleic acid-binding OB-folds from interaction proteins A3 and A6 that surprisingly, form a heterodimer. Crystal growth required the assistance of an anti-A3 nanobody as a crystallization chaperone. Unexpectedly, this anti-A3 nanobody binds to both A3(OB) and A6, despite only ~40% amino acid sequence identity between the OB-folds of A3 and A6. The A3(OB)-A6 heterodimer buries 35% more surface area than the A6 homodimer. This is attributed mainly to the presence of a conserved Pro-rich loop in A3(OB). The implications of the A3(OB)-A6 heterodimer, and of a dimer of heterodimers observed in the crystals, for the architecture of the editosome are profound, resulting in a proposal of a 'five OB-fold center' in the core of the editosome.
Organizational Affiliation:
Department of Biochemistry, Biomolecular Structure Center, School of Medicine, University of Washington, PO Box 357742, Seattle WA 98195, USA.