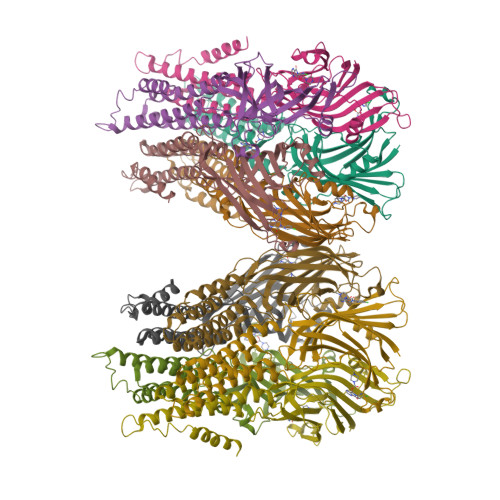
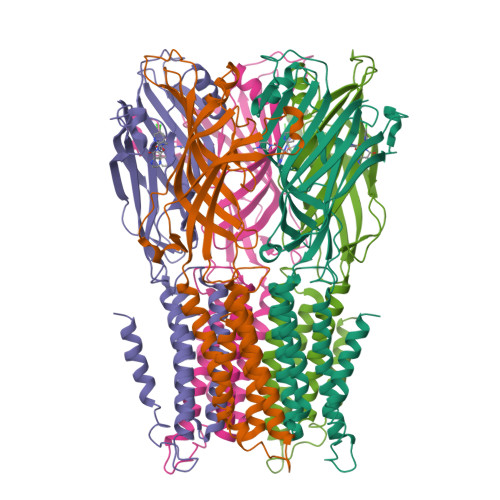
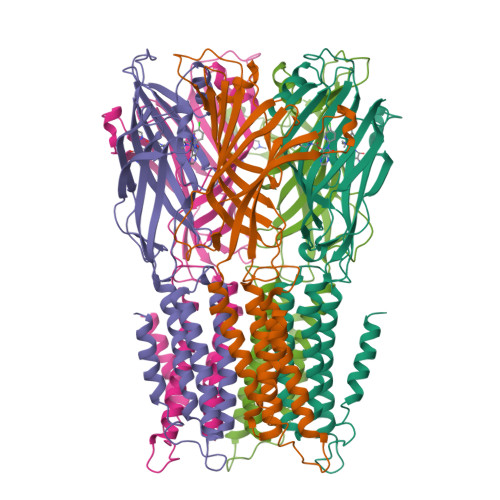
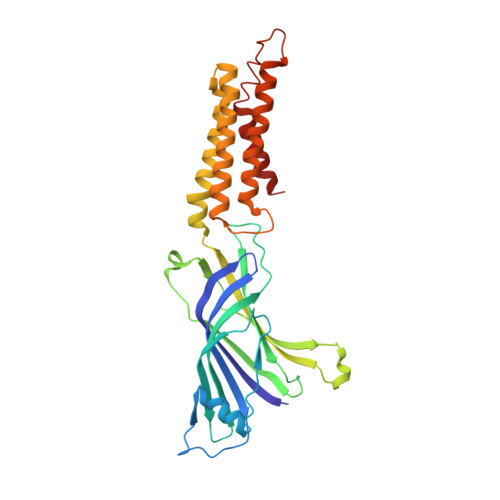
Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Spurny, R., Ramerstorfer, J., Price, K., Brams, M., Ernst, M., Nury, H., Verheij, M., Legrand, P., Bertrand, D., Bertrand, S., Dougherty, D.A., De Esch, I.J.P., Corringer, P., Sieghart, W., Lummis, S.C.R., Ulens, C.(2012) Proc Natl Acad Sci U S A 109: E3028
- PubMed: 23035248
- DOI: https://doi.org/10.1073/pnas.1208208109
- Primary Citation of Related Structures:
2YOE, 4A97, 4A98 - PubMed Abstract:
GABA(A) receptors are pentameric ligand-gated ion channels involved in fast inhibitory neurotransmission and are allosterically modulated by the anxiolytic, anticonvulsant, and sedative-hypnotic benzodiazepines. Here we show that the prokaryotic homolog ELIC also is activated by GABA and is modulated by benzodiazepines with effects comparable to those at GABA(A) receptors. Crystal structures reveal important features of GABA recognition and indicate that benzodiazepines, depending on their concentration, occupy two possible sites in ELIC. An intrasubunit site is adjacent to the GABA-recognition site but faces the channel vestibule. A second intersubunit site partially overlaps with the GABA site and likely corresponds to a low-affinity benzodiazepine-binding site in GABA(A) receptors that mediates inhibitory effects of the benzodiazepine flurazepam. Our study offers a structural view how GABA and benzodiazepines are recognized at a GABA-activated ion channel.
Organizational Affiliation:
Department of Cellular and Molecular Medicine, Laboratory of Structural Neurobiology, Catholic University of Leuven, 3000 Leuven, Belgium.