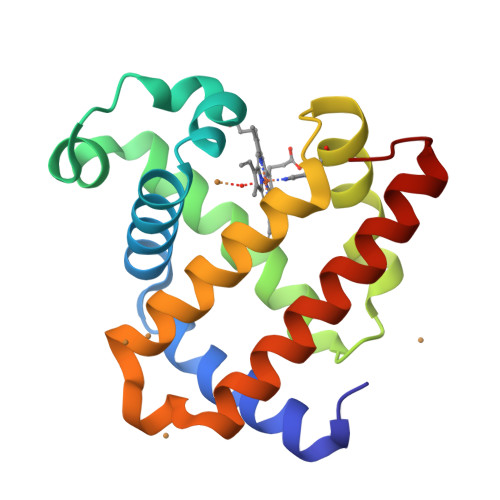
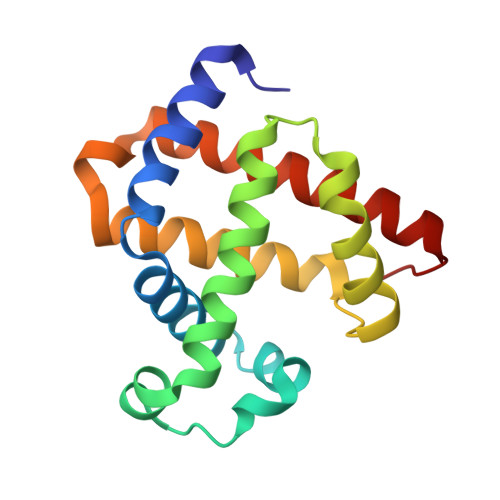
A Designed Functional Metalloenzyme that Reduces O(2) to H(2) O with Over One Thousand Turnovers.
Miner, K.D., Mukherjee, A., Gao, Y.G., Null, E.L., Petrik, I.D., Zhao, X., Yeung, N., Robinson, H., Lu, Y.(2012) Angew Chem Int Ed Engl 51: 5589-5592
- PubMed: 22539151
- DOI: https://doi.org/10.1002/anie.201201981
- Primary Citation of Related Structures:
4FWX, 4FWY, 4FWZ - PubMed Abstract:
[Image: see text] Rational design of functional enzymes with high turnovers is a significant challenge, especially those with complex active site and difficult reactions, such as in respiratory oxidases. Introducing 2 His and 1 Tyr into myoglobin resulted in designed enzymes that reduce O 2 to H 2 O with > 1000 turnovers and minimal release of reactive oxygen species. This also showed that presence and positioning of Tyr, not Cu, are critical for activity.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA.