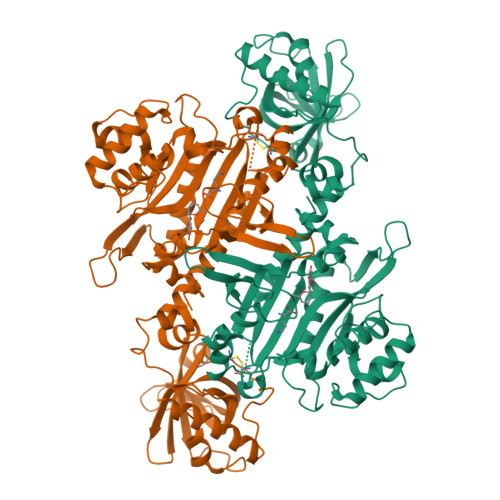
Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
Khan, S., Sharma, A., Belrhali, H., Yogavel, M., Sharma, A.(2014) J Struct Funct Genomics 15: 63-71
- PubMed: 24935905
- DOI: https://doi.org/10.1007/s10969-014-9182-1
- Primary Citation of Related Structures:
4PG3 - PubMed Abstract:
Malaria parasites inevitably develop drug resistance to anti-malarials over time. Hence the immediacy for discovering new chemical scaffolds to include in combination malaria drug therapy. The desirable attributes of new chemotherapeutic agents currently include activity against both liver and blood stage malaria parasites. One such recently discovered compound called cladosporin abrogates parasite growth via inhibition of Plasmodium falciparum lysyl-tRNA synthetase (PfKRS), an enzyme central to protein translation. Here, we present crystal structure of ternary PfKRS-lysine-cladosporin (PfKRS-K-C) complex that reveals cladosporin's remarkable ability to mimic the natural substrate adenosine and thereby colonize PfKRS active site. The isocoumarin fragment of cladosporin sandwiches between critical adenine-recognizing residues while its pyran ring fits snugly in the ribose-recognizing cavity. PfKRS-K-C structure highlights ample space within PfKRS active site for further chemical derivatization of cladosporin. Such derivatives may be useful against additional human pathogens that retain high conservation in cladosporin chelating residues within their lysyl-tRNA synthetase.
Organizational Affiliation:
Structural and Computational Biology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, 110067, India.