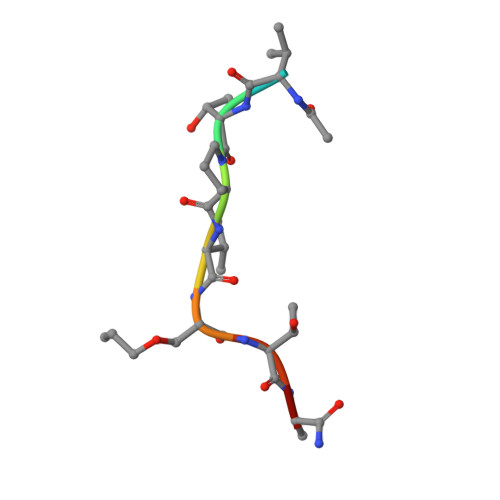
Bisubstrate Udp-Peptide Conjugates as Human O-Glcnac Transferase Inhibitors.
Borodkin, V.S., Schimpl, M., Gundogdu, M., Rafie, K., Dorfmueller, H.C., Robinson, D.A., Van Aalten, D.M.(2014) Biochem J 457: 497
- PubMed: 24256146
- DOI: https://doi.org/10.1042/BJ20131272
- Primary Citation of Related Structures:
4CDR - PubMed Abstract:
Inhibitors of OGT (O-GlcNAc transferase) are valuable tools to study the cell biology of protein O-GlcNAcylation. We report OGT bisubstrate-linked inhibitors (goblins) in which the acceptor serine in the peptide VTPVSTA is covalently linked to UDP, eliminating the GlcNAc pyranoside ring. Goblin1 co-crystallizes with OGT, revealing an ordered C₃ linker and retained substrate-binding modes, and binds the enzyme with micromolar affinity, inhibiting glycosyltransfer on to protein and peptide substrates.
Organizational Affiliation:
*MRC Protein Phosphorylation und Ubiquitylation Unit, College of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, U.K.