Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Martinez-Saez, N., Castro-Lopez, J., Valero-Gonzalez, J., Madariaga, D., Companon, I., Somovilla, V.J., Salvado, M., Asensio, J.L., Jimenez-Barbero, J., Avenoza, A., Busto, J.H., Bernardes, G.J.L., Peregrina, J.M., Hurtado-Guerrero, R., Corzana, F.(2015) Angew Chem Int Ed Engl 54: 9830
- PubMed: 26118689
- DOI: https://doi.org/10.1002/anie.201502813
- Primary Citation of Related Structures:
5A2I, 5A2J, 5A2K, 5A2L - PubMed Abstract:
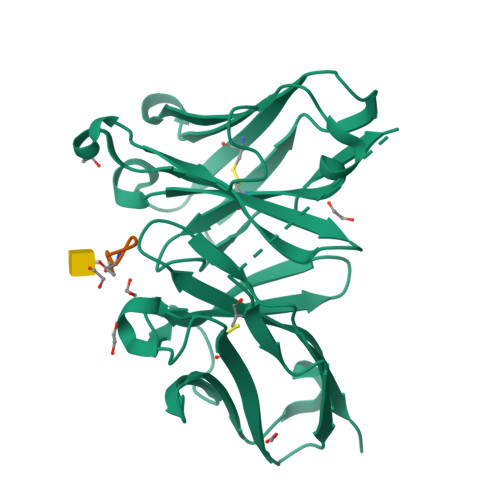
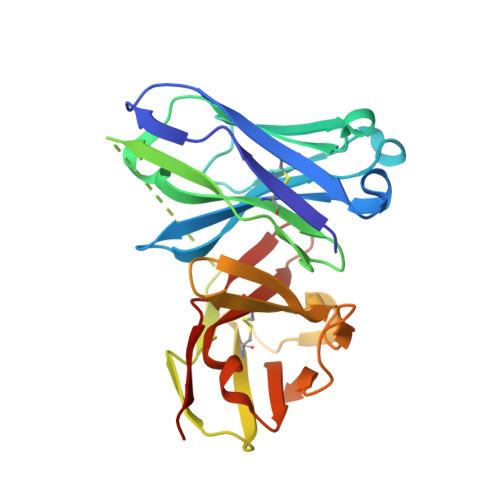
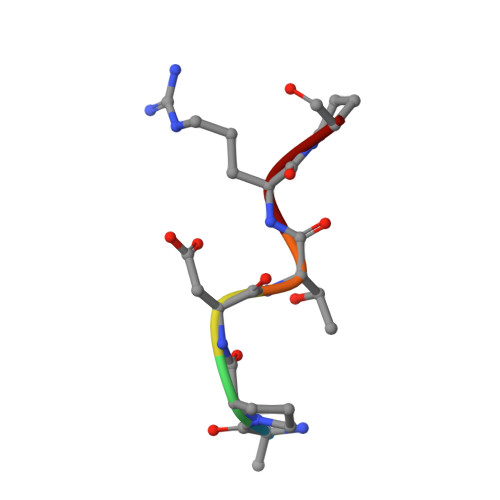
The structural features of MUC1-like glycopeptides bearing the Tn antigen (α-O-GalNAc-Ser/Thr) in complex with an anti MUC-1 antibody are reported at atomic resolution. For the α-O-GalNAc-Ser derivative, the glycosidic linkage adopts a high-energy conformation, barely populated in the free state. This unusual structure (also observed in an α-S-GalNAc-Cys mimic) is stabilized by hydrogen bonds between the peptidic fragment and the sugar. The selection of a particular peptide structure by the antibody is thus propagated to the carbohydrate through carbohydrate/peptide contacts, which force a change in the orientation of the sugar moiety. This seems to be unfeasible in the α-O-GalNAc-Thr glycopeptide owing to the more limited flexibility of the side chain imposed by the methyl group. Our data demonstrate the non-equivalence of Ser and Thr O-glycosylation points in molecular recognition processes. These features provide insight into the occurrence in nature of the APDTRP epitope for anti-MUC1 antibodies.
Organizational Affiliation:
Departamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química, 26006 Logroño (Spain).