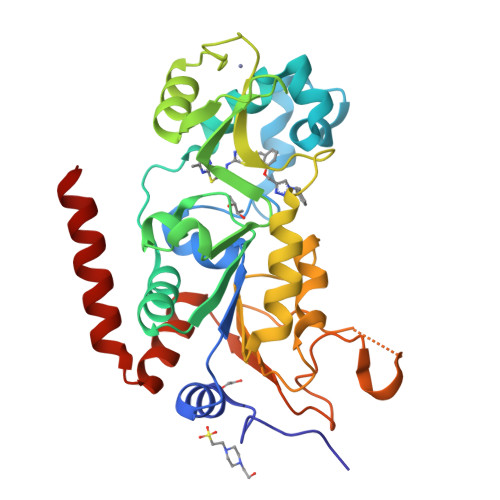
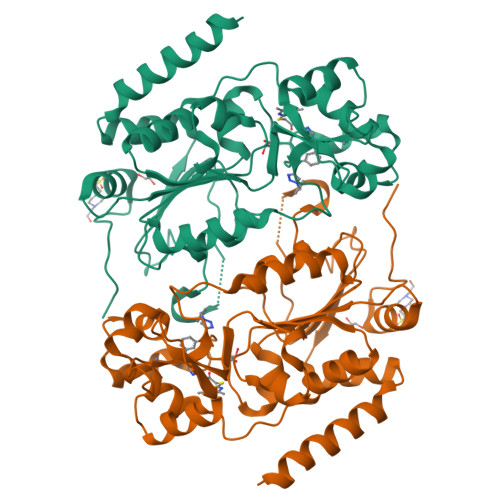
Structure-Based Development of an Affinity Probe for Sirtuin 2.
Schiedel, M., Rumpf, T., Karaman, B., Lehotzky, A., Gerhardt, S., Ovadi, J., Sippl, W., Einsle, O., Jung, M.(2016) Angew Chem Int Ed Engl 55: 2252-2256
- PubMed: 26748890
- DOI: https://doi.org/10.1002/anie.201509843
- Primary Citation of Related Structures:
5DY5 - PubMed Abstract:
Sirtuins are NAD(+)-dependent protein deacylases that cleave off acetyl groups, as well as other acyl groups, from the ɛ-amino group of lysines in histones and other substrate proteins. Dysregulation of human Sirt2 activity has been associated with the pathogenesis of cancer, inflammation, and neurodegeneration, thus making Sirt2 a promising target for pharmaceutical intervention. Here, based on a crystal structure of Sirt2 in complex with an optimized sirtuin rearranging ligand (SirReal) that shows improved potency, water solubility, and cellular efficacy, we present the development of the first Sirt2-selective affinity probe. A slow dissociation of the probe/enzyme complex offers new applications for SirReals, such as biophysical characterization, fragment-based screening, and affinity pull-down assays. This possibility makes the SirReal probe an important tool for studying sirtuin biology.
Organizational Affiliation:
Institut für Pharmazeutische Wissenschaften, Albert-Ludwigs-Universität Freiburg, Albertstrasse 25, 79104, Freiburg im Breisgau, Germany.