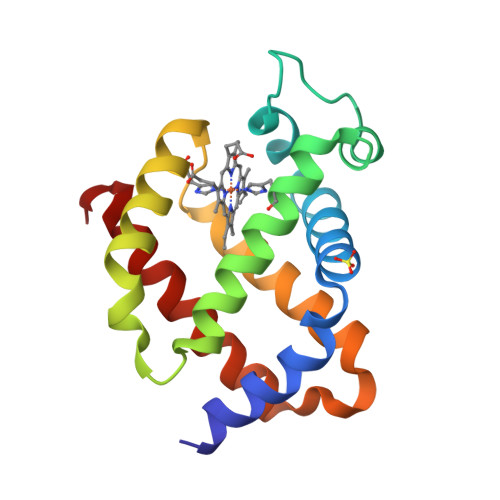
Determinants of neuroglobin plasticity highlighted by joint coarse-grained simulations and high pressure crystallography.
Colloc'h, N., Sacquin-Mora, S., Avella, G., Dhaussy, A.C., Prange, T., Vallone, B., Girard, E.(2017) Sci Rep 7: 1858-1858
- PubMed: 28500341
- DOI: https://doi.org/10.1038/s41598-017-02097-1
- Primary Citation of Related Structures:
5EET, 5EOH, 5EQM, 5EU2, 5EV5, 5EYJ, 5EYS, 5F0B, 5F2A - PubMed Abstract:
Investigating the effect of pressure sheds light on the dynamics and plasticity of proteins, intrinsically correlated to functional efficiency. Here we detail the structural response to pressure of neuroglobin (Ngb), a hexacoordinate globin likely to be involved in neuroprotection. In murine Ngb, reversible coordination is achieved by repositioning the heme more deeply into a large internal cavity, the "heme sliding mechanism". Combining high pressure crystallography and coarse-grain simulations on wild type Ngb as well as two mutants, one (V101F) with unaffected and another (F106W) with decreased affinity for CO, we show that Ngb hinges around a rigid mechanical nucleus of five hydrophobic residues (V68, I72, V109, L113, Y137) during its conformational transition induced by gaseous ligand, that the intrinsic flexibility of the F-G loop appears essential to drive the heme sliding mechanism, and that residue Val 101 may act as a sensor of the interaction disruption between the heme and the distal histidine.
Organizational Affiliation:
ISTCT CNRS UNICAEN CEA Normandie Univ., CERVOxy team, centre Cyceron, 14000, Caen, France. colloch@cyceron.fr.