SERK Family Receptor-like Kinases Function as Co-receptors with PXY for Plant Vascular Development
Zhang, H.Q., Lin, X.Y., Han, Z.F., Wang, J., Qu, L.J., Chai, J.J.(2016) Mol Plant 9: 1406-1414
- PubMed: 27449136
- DOI: https://doi.org/10.1016/j.molp.2016.07.004
- Primary Citation of Related Structures:
5GQR - PubMed Abstract:
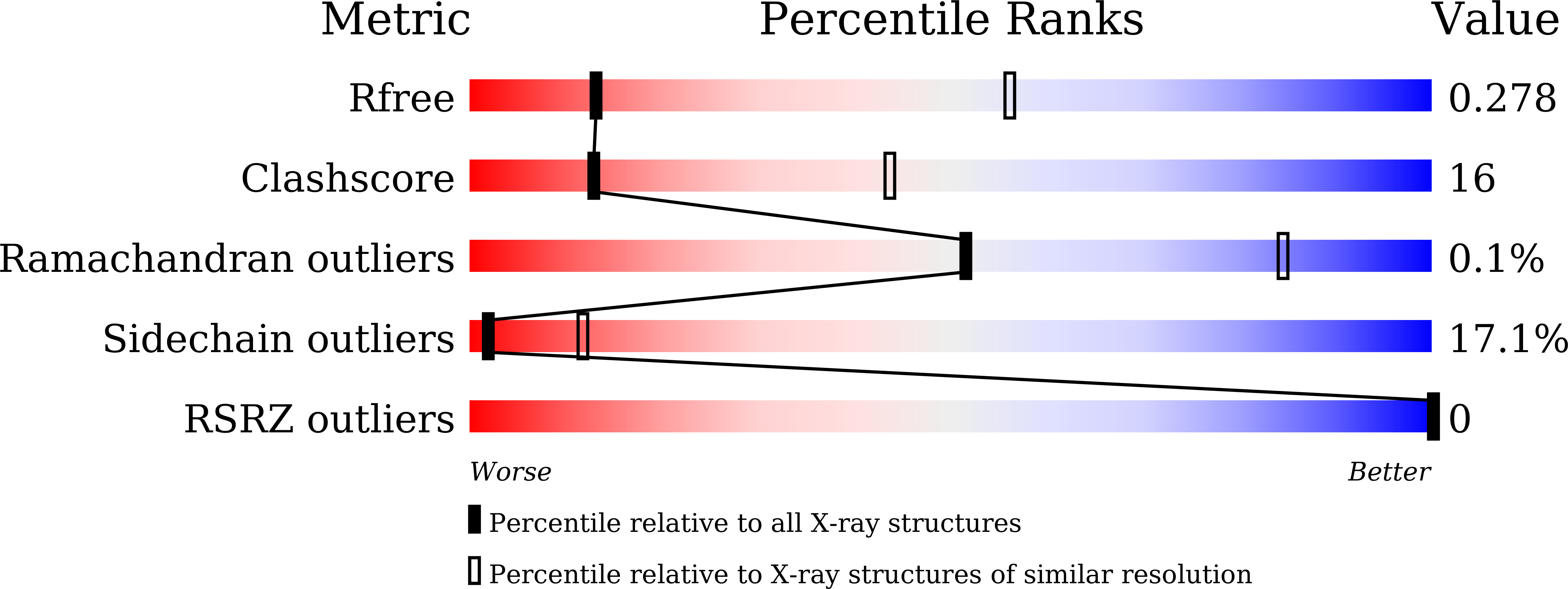
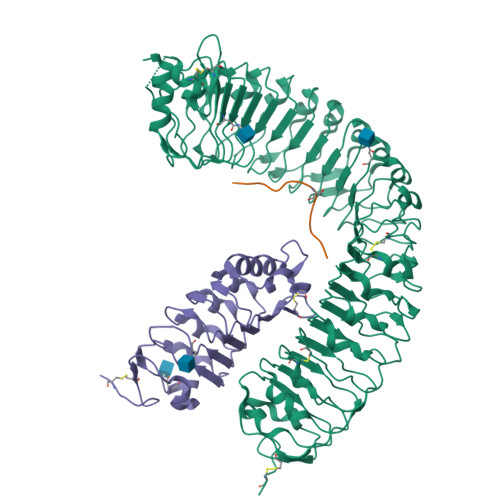
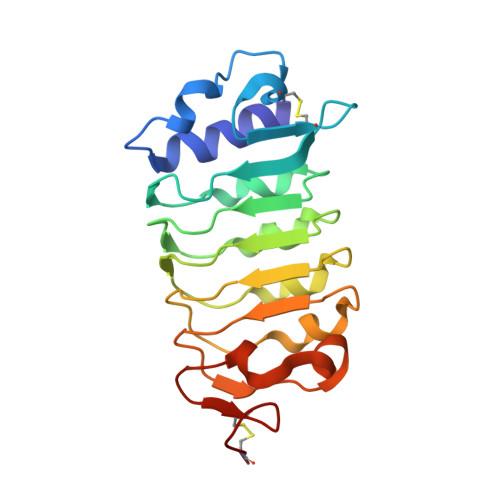
In Arabidopsis, the CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptides play important roles in regulating proliferation and differentiation of plant-specific stem cells. Although receptors of CLEs are reported to be leucine-rich repeat receptor kinases, the mechanisms underlying CLE-induced receptor activation remain largely unknown. Here we show that SOMATIC EMBRYOGENESIS RECEPTOR KINASEs (SERKs) serve as co-receptors in CLE41/TDIF-PXY signaling to regulate plant vascular development. TDIF induces interaction of its receptor PXY with SERKs in vitro and in vivo. Furthermore, the serk1-1 serk2-1 bak1-5 mutant plants are less sensitive to TDIF, phenocopying the pxy mutant with a compromised promotion of procambial cell proliferation. Crystal structure of the PXY-TDIF-SERK2 complex reveals that the last amino acid of TDIF conserved among CLEs and other evolutionary-related peptides is important for the interaction between SERK2 and PXY. Taken together, our current study identifies SERKs as signaling components of the TDIF-PXY pathway and suggests a conserved activation mechanism of CLE receptors.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China.