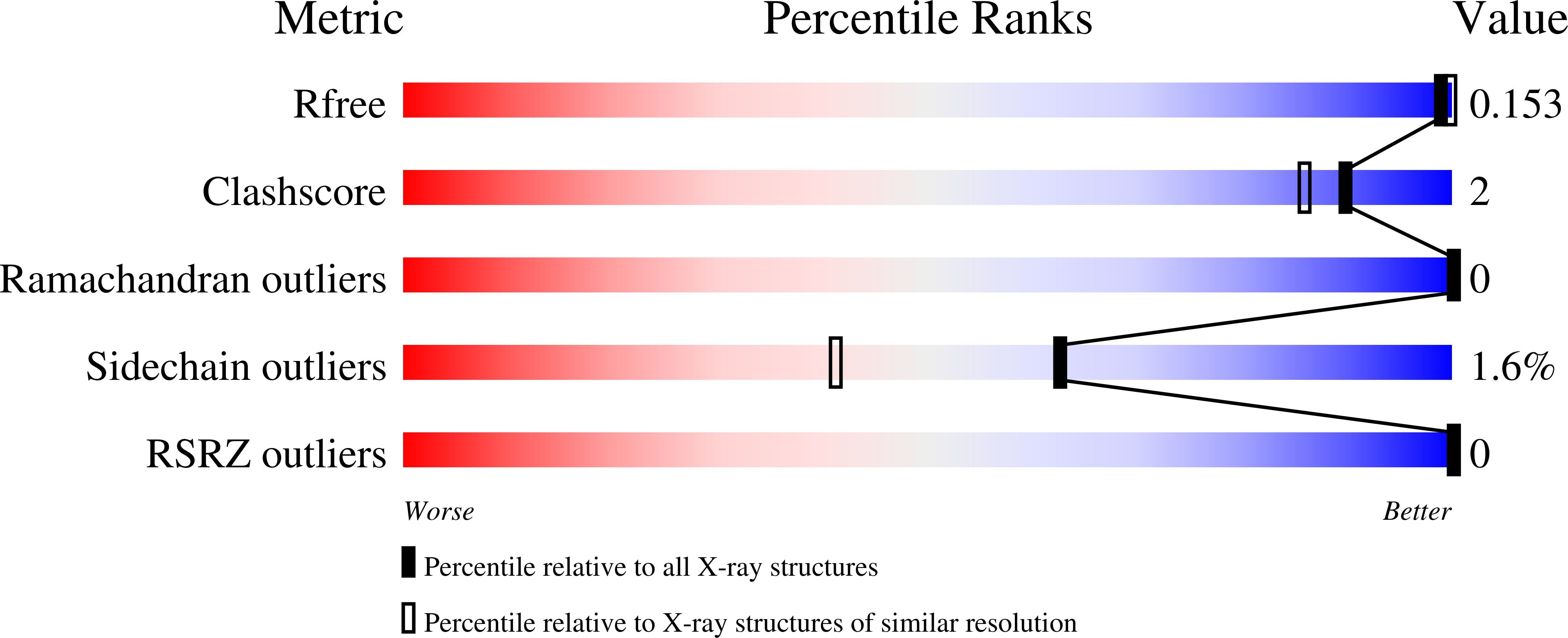
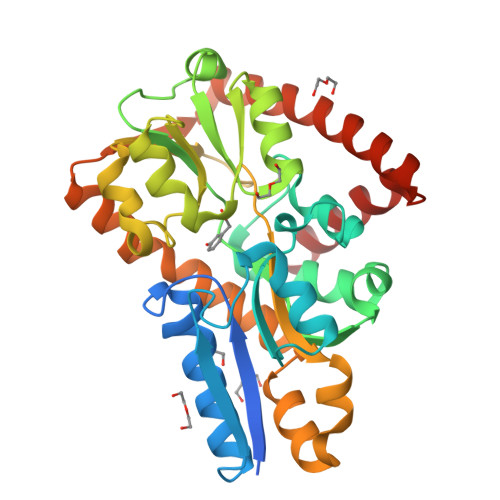
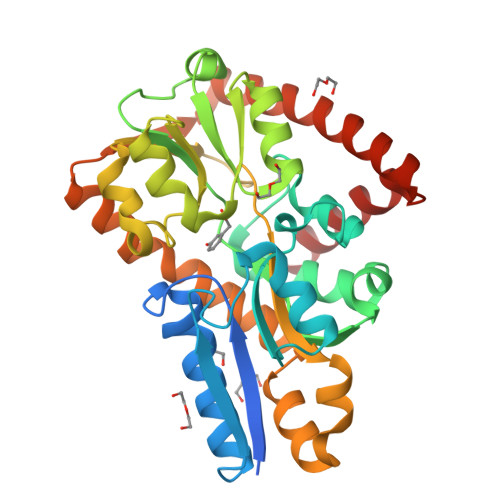
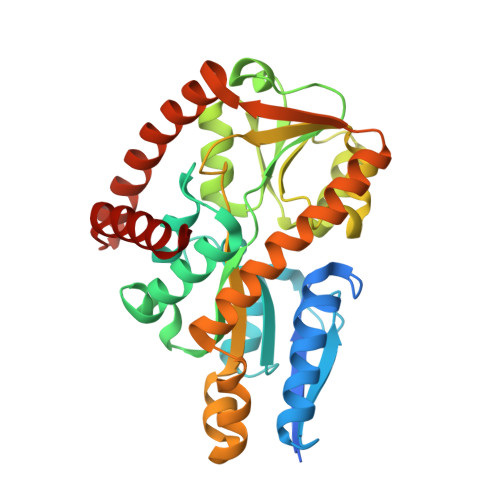
Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
Vetting, M.W., Al Obaidi, N.F., Hogle, S.L., Dupont, C.L., Almo, S.C.To be published.