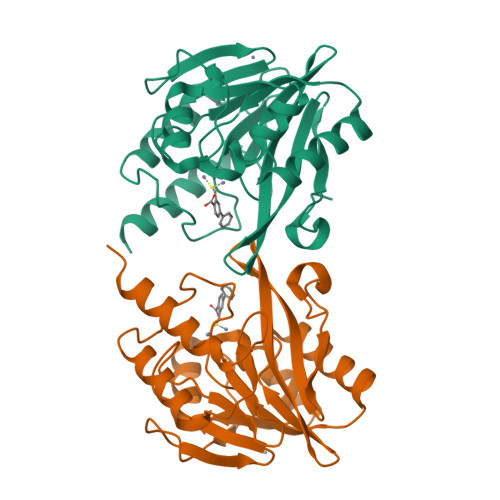
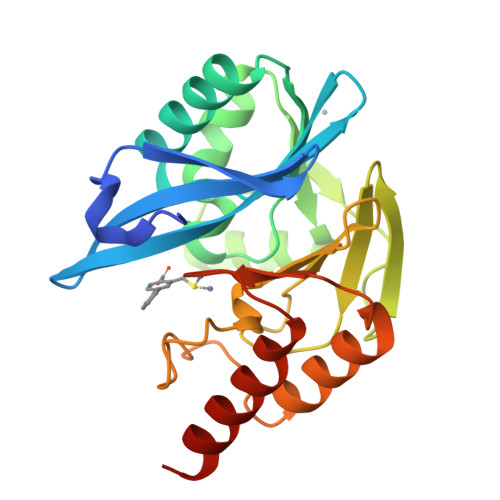
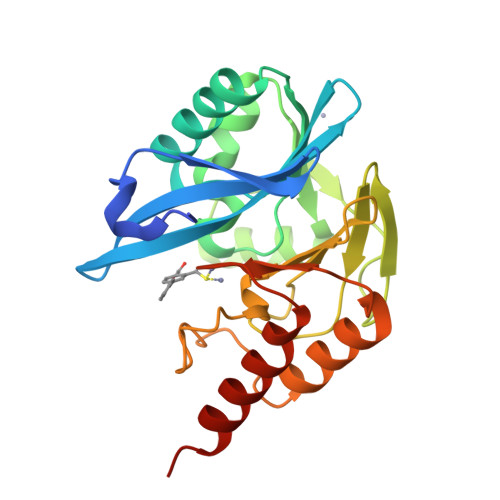
In Silico Fragment-Based Design Identifies Subfamily B1 Metallo-beta-lactamase Inhibitors.
Cain, R., Brem, J., Zollman, D., McDonough, M.A., Johnson, R.M., Spencer, J., Makena, A., Abboud, M.I., Cahill, S., Lee, S.Y., McHugh, P.J., Schofield, C.J., Fishwick, C.W.G.(2018) J Med Chem 61: 1255-1260
- PubMed: 29271657
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01728
- Primary Citation of Related Structures:
5K48 - PubMed Abstract:
Zinc ion-dependent β-lactamases (MBLs) catalyze the hydrolysis of almost all β-lactam antibiotics and resist the action of clinically available β-lactamase inhibitors. We report how application of in silico fragment-based molecular design employing thiol-mediated metal anchorage leads to potent MBL inhibitors. The new inhibitors manifest potent inhibition of clinically important B1 subfamily MBLs, including the widespread NDM-1, IMP-1, and VIM-2 enzymes; with lower potency, some of them also inhibit clinically relevant Class A and D serine-β-lactamases. The inhibitors show selectivity for bacterial MBL enzymes compared to that for human MBL fold nucleases. Cocrystallization of one inhibitor, which shows potentiation of Meropenem activity against MBL-expressing Enterobacteriaceae, with VIM-2 reveals an unexpected binding mode, involving interactions with residues from conserved active site bordering loops.
Organizational Affiliation:
School of Chemistry, University of Leeds , Leeds LS2 9JT, United Kingdom.