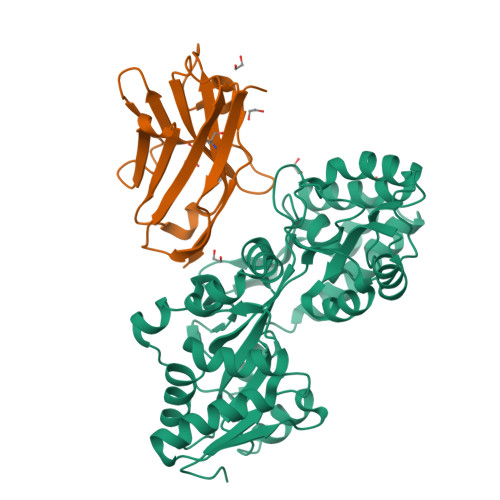
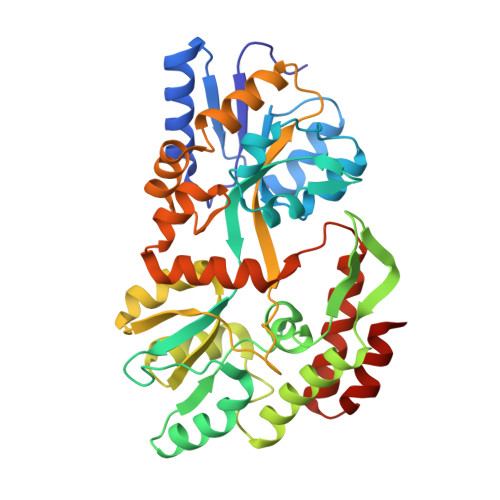
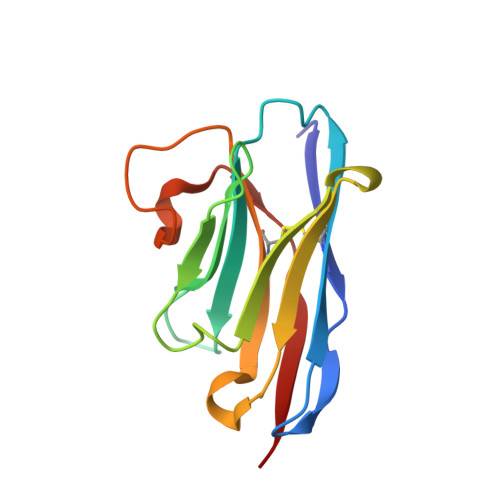
Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Zimmermann, I., Egloff, P., Hutter, C.A., Arnold, F.M., Stohler, P., Bocquet, N., Hug, M.N., Huber, S., Siegrist, M., Hetemann, L., Gera, J., Gmur, S., Spies, P., Gygax, D., Geertsma, E.R., Dawson, R.J., Seeger, M.A.(2018) Elife 7
- PubMed: 29792401
- DOI: https://doi.org/10.7554/eLife.34317
- Primary Citation of Related Structures:
5M13, 5M14, 5M15 - PubMed Abstract:
Mechanistic and structural studies of membrane proteins require their stabilization in specific conformations. Single domain antibodies are potent reagents for this purpose, but their generation relies on immunizations, which impedes selections in the presence of ligands typically needed to populate defined conformational states. To overcome this key limitation, we developed an in vitro selection platform based on synthetic single domain antibodies named sybodies. To target the limited hydrophilic surfaces of membrane proteins, we designed three sybody libraries that exhibit different shapes and moderate hydrophobicity of the randomized surface. A robust binder selection cascade combining ribosome and phage display enabled the generation of conformation-selective, high affinity sybodies against an ABC transporter and two previously intractable human SLC transporters, GlyT1 and ENT1. The platform does not require access to animal facilities and builds exclusively on commercially available reagents, thus enabling every lab to rapidly generate binders against challenging membrane proteins.
Organizational Affiliation:
Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.