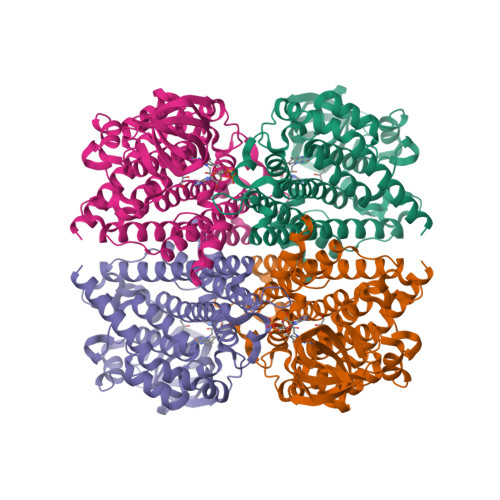
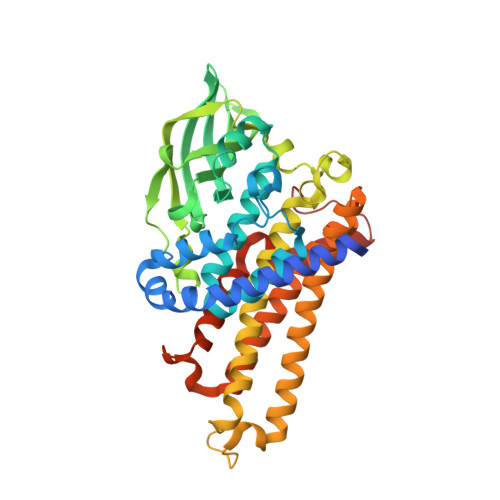
Cryptic indole hydroxylation by a non-canonical terpenoid cyclase parallels bacterial xenobiotic detoxification.
Kugel, S., Baunach, M., Baer, P., Ishida-Ito, M., Sundaram, S., Xu, Z., Groll, M., Hertweck, C.(2017) Nat Commun 8: 15804-15804
- PubMed: 28643772
- DOI: https://doi.org/10.1038/ncomms15804
- Primary Citation of Related Structures:
5LVU, 5LVW, 5MR6 - PubMed Abstract:
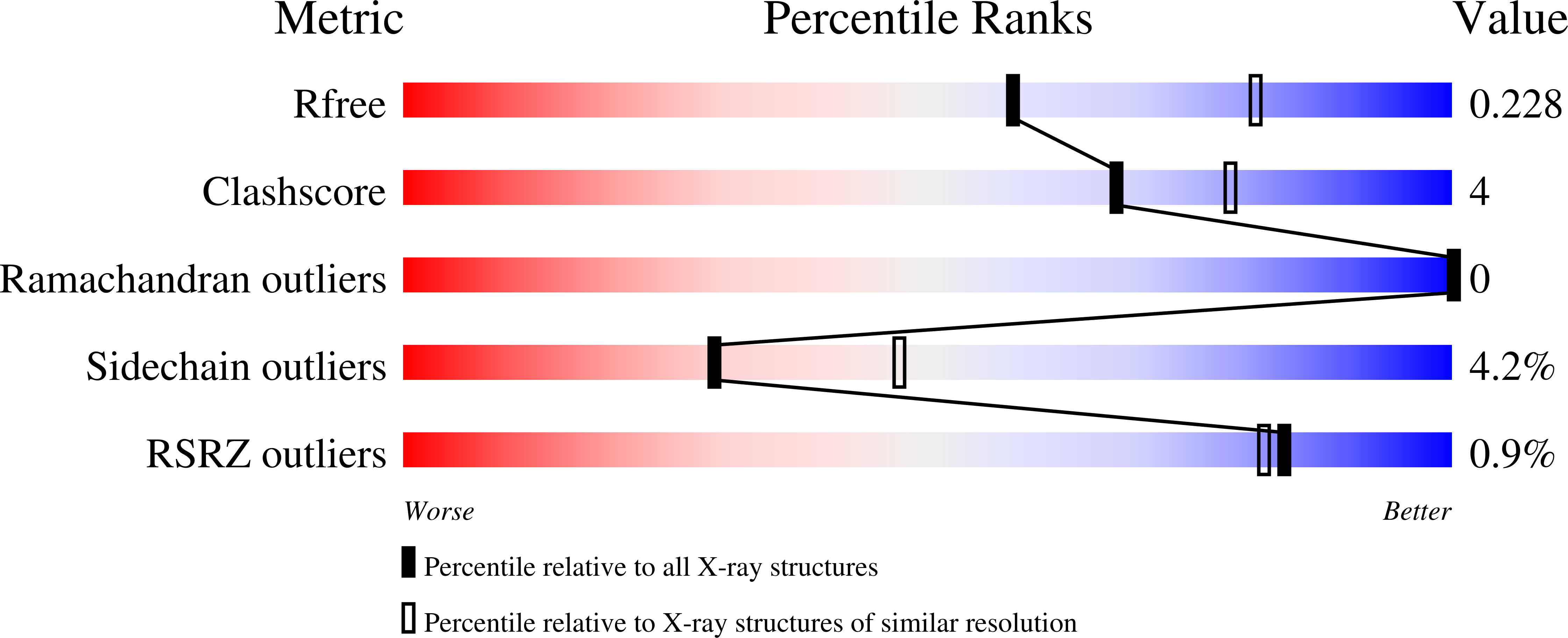
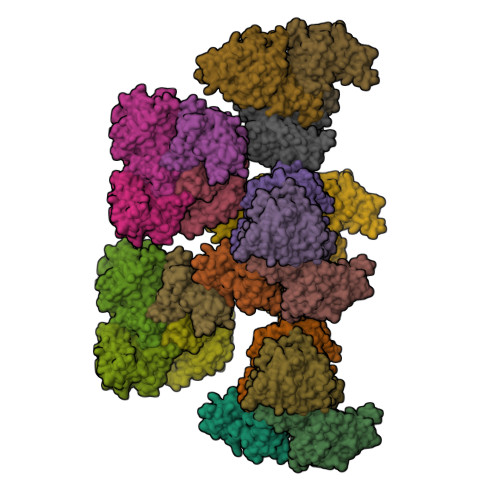
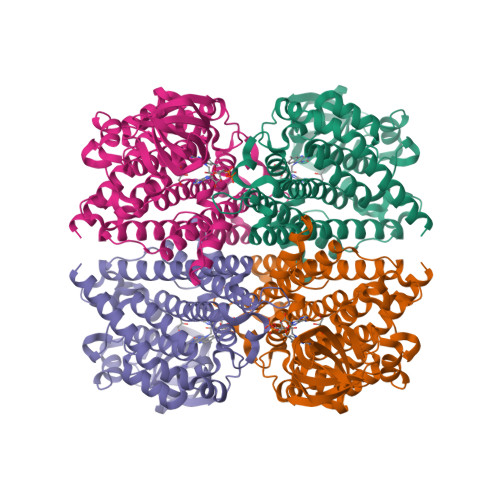
Terpenoid natural products comprise a wide range of molecular architectures that typically result from C-C bond formations catalysed by classical type I/II terpene cyclases. However, the molecular diversity of biologically active terpenoids is substantially increased by fully unrelated, non-canonical terpenoid cyclases. Their evolutionary origin has remained enigmatic. Here we report the in vitro reconstitution of an unusual flavin-dependent bacterial indoloterpenoid cyclase, XiaF, together with a designated flavoenzyme-reductase (XiaP) that mediates a key step in xiamycin biosynthesis. The crystal structure of XiaF with bound FADH 2 (at 2.4 Å resolution) and phylogenetic analyses reveal that XiaF is, surprisingly, most closely related to xenobiotic-degrading enzymes. Biotransformation assays show that XiaF is a designated indole hydroxylase that can be used for the production of indigo and indirubin. We unveil a cryptic hydroxylation step that sets the basis for terpenoid cyclization and suggest that the cyclase has evolved from xenobiotics detoxification enzymes.
Organizational Affiliation:
Department of Biomolecular Chemistry, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany.