Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
Douzi, B., Trinh, N.T.T., Michel-Souzy, S., Desmyter, A., Ball, G., Barbier, P., Kosta, A., Durand, E., Forest, K.T., Cambillau, C., Roussel, A., Voulhoux, R.(2017) mBio 8
- PubMed: 29042493
- DOI: https://doi.org/10.1128/mBio.01185-17
- Primary Citation of Related Structures:
5MP2, 5NGI - PubMed Abstract:
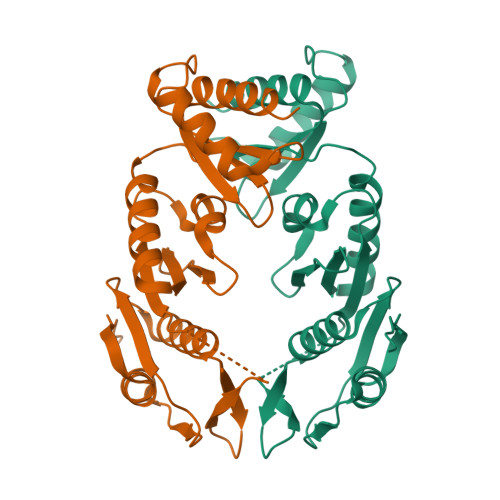
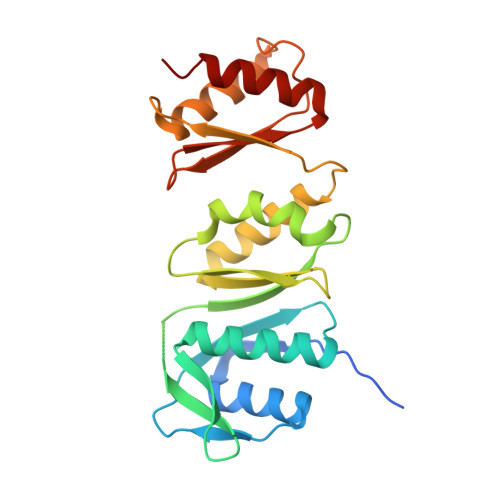
The type II secretion system (T2SS) releases large folded exoproteins across the envelope of many Gram-negative pathogens. This secretion process therefore requires specific gating, interacting, and dynamics properties mainly operated by a bipartite outer membrane channel called secretin. We have a good understanding of the structure-function relationship of the pore-forming C-terminal domain of secretins. In contrast, the high flexibility of their periplasmic N-terminal domain has been an obstacle in obtaining the detailed structural information required to uncover its molecular function. In Pseudomonas aeruginosa , the Xcp T2SS plays an important role in bacterial virulence by its capacity to deliver a large panel of toxins and degradative enzymes into the surrounding environment. Here, we revealed that the N-terminal domain of XcpQ secretin spontaneously self-assembled into a hexamer of dimers independently of its C-terminal domain. Furthermore, and by using multidisciplinary approaches, we elucidate the structural organization of the XcpQ N domain and demonstrate that secretin flexibility at interdimer interfaces is mandatory for its function. IMPORTANCE Bacterial secretins are large homooligomeric proteins constituting the outer membrane pore-forming element of several envelope-embedded nanomachines essential in bacterial survival and pathogenicity. They comprise a well-defined membrane-embedded C-terminal domain and a modular periplasmic N-terminal domain involved in substrate recruitment and connection with inner membrane components. We are studying the XcpQ secretin of the T2SS present in the pathogenic bacterium Pseudomonas aeruginosa Our data highlight the ability of the XcpQ N-terminal domain to spontaneously oligomerize into a hexamer of dimers. Further in vivo experiments revealed that this domain adopts different conformations essential for the T2SS secretion process. These findings provide new insights into the functional understanding of bacterial T2SS secretins.
Organizational Affiliation:
Aix Marseille University, CNRS, IMM, LISM, Marseille, France bdouzi@imm.cnrs.fr voulhoux@imm.cnrs.fr.