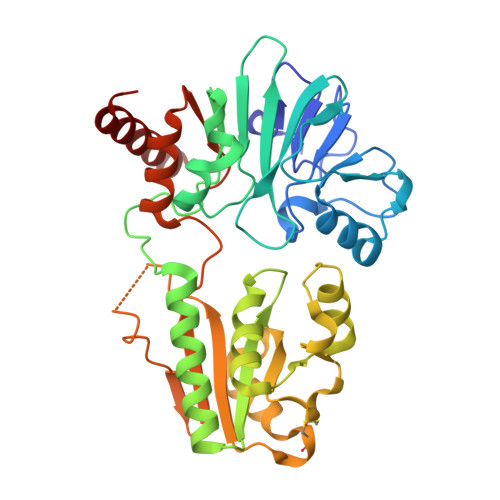
Crystal structure of DNA cross-link repair protein 1A in complex with Ceftriaxone (alternative site)
Newman, J.A., Aitkenhead, H., Kupinska, K., Burgess-Brown, N.A., Talon, R., Krojer, T., von Delft, F., Arrowsmith, C.H., Edwards, A., Bountra, C., Gileadi, O.To be published.