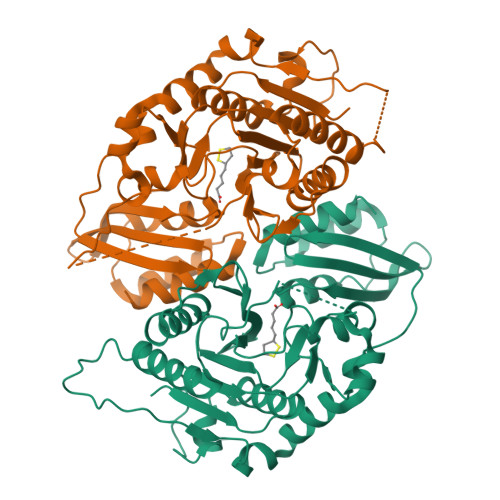
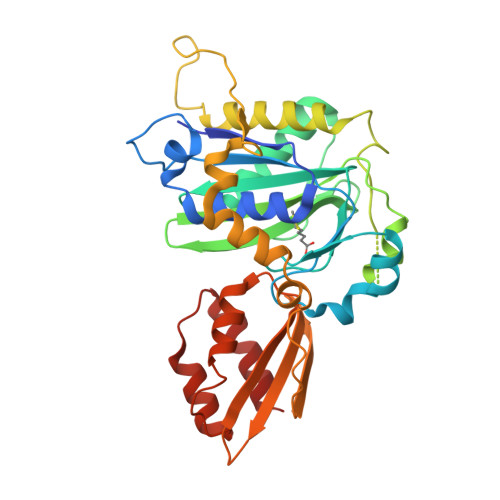
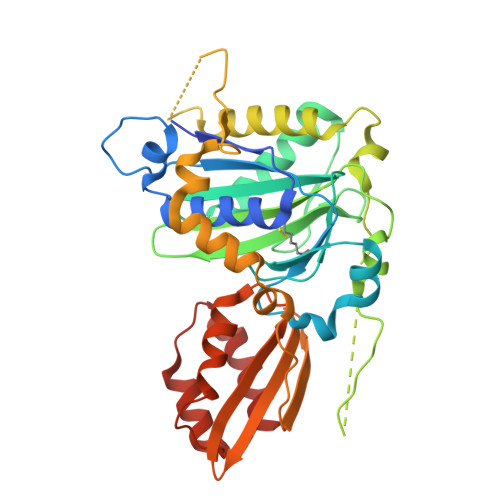
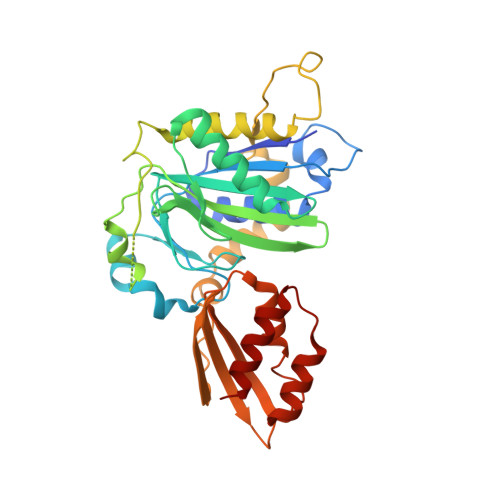
Crystal structure of lipoate-bound lipoate ligase 1, LipL1, from Plasmodium falciparum.
Guerra, A.J., Afanador, G.A., Prigge, S.T.(2017) Proteins 85: 1777-1783
- PubMed: 28543853
- DOI: https://doi.org/10.1002/prot.25324
- Primary Citation of Related Structures:
5T8U - PubMed Abstract:
Plasmodium falciparum lipoate protein ligase 1 (PfLipL1) is an ATP-dependent ligase that belongs to the biotin/lipoate A/B protein ligase family (PFAM PF03099). PfLipL1 is the only known canonical lipoate ligase in Pf and functions as a redox switch between two lipoylation routes in the parasite mitochondrion. Here, we report the crystal structure of a deletion construct of PfLipL1 (PfLipL1 Δ243-279 ) bound to lipoate, and validate the lipoylation activity of this construct in both an in vitro lipoylation assay and a cell-based lipoylation assay. This characterization represents the first step in understanding the redox dependence of the lipoylation mechanism in malaria parasites. Proteins 2017; 85:1777-1783. © 2017 Wiley Periodicals, Inc.
Organizational Affiliation:
Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland.