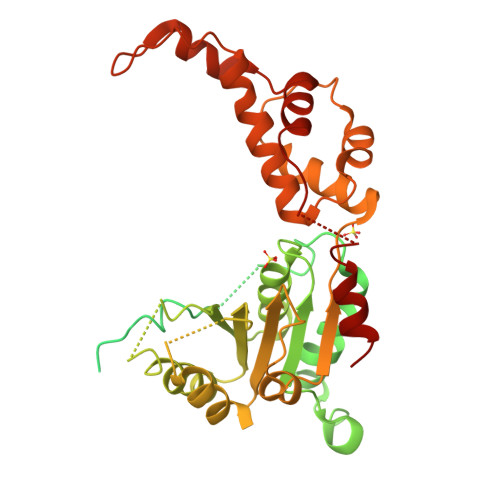
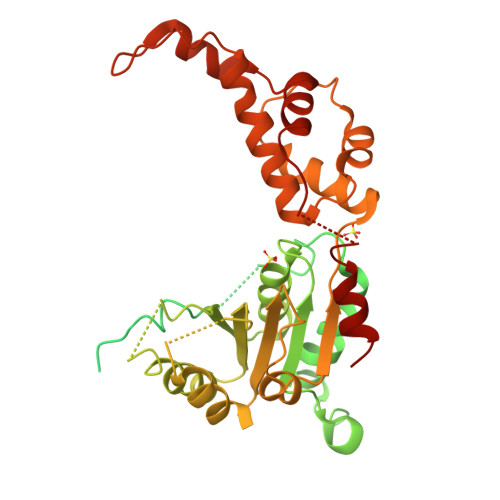
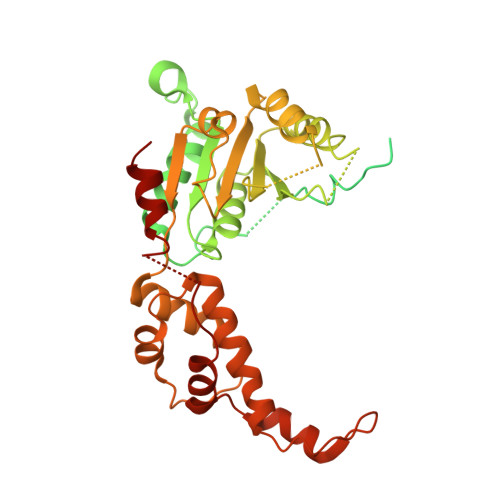
Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Zehr, E., Szyk, A., Piszczek, G., Szczesna, E., Zuo, X., Roll-Mecak, A.(2017) Nat Struct Mol Biol 24: 717-725
- PubMed: 28783150
- DOI: https://doi.org/10.1038/nsmb.3448
- Primary Citation of Related Structures:
5WC0, 5WC1, 5WCB - PubMed Abstract:
Microtubule-severing enzymes katanin, spastin and fidgetin are AAA ATPases important for the biogenesis and maintenance of complex microtubule arrays in axons, spindles and cilia. Because of a lack of known 3D structures for these enzymes, their mechanism of action has remained poorly understood. Here we report the X-ray crystal structure of the monomeric AAA katanin module from Caenorhabditis elegans and cryo-EM reconstructions of the hexamer in two conformations. The structures reveal an unexpected asymmetric arrangement of the AAA domains mediated by structural elements unique to microtubule-severing enzymes and critical for their function. The reconstructions show that katanin cycles between open spiral and closed ring conformations, depending on the ATP occupancy of a gating protomer that tenses or relaxes interprotomer interfaces. Cycling of the hexamer between these conformations would provide the power stroke for microtubule severing.
Organizational Affiliation:
Cell Biology and Biophysics Unit, Porter Neuroscience Research Center, National Institute of Neurological Disorders and Stroke, Bethesda, Maryland, USA.