Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Liao, H.J., Li, J., Huang, J.L., Davidson, M., Kurnikov, I., Lin, T.S., Lee, J.L., Kurnikova, M., Guo, Y., Chan, N.L., Chang, W.C.(2018) Angew Chem Int Ed Engl 57: 1831-1835
- PubMed: 29314482
- DOI: https://doi.org/10.1002/anie.201710567
- Primary Citation of Related Structures:
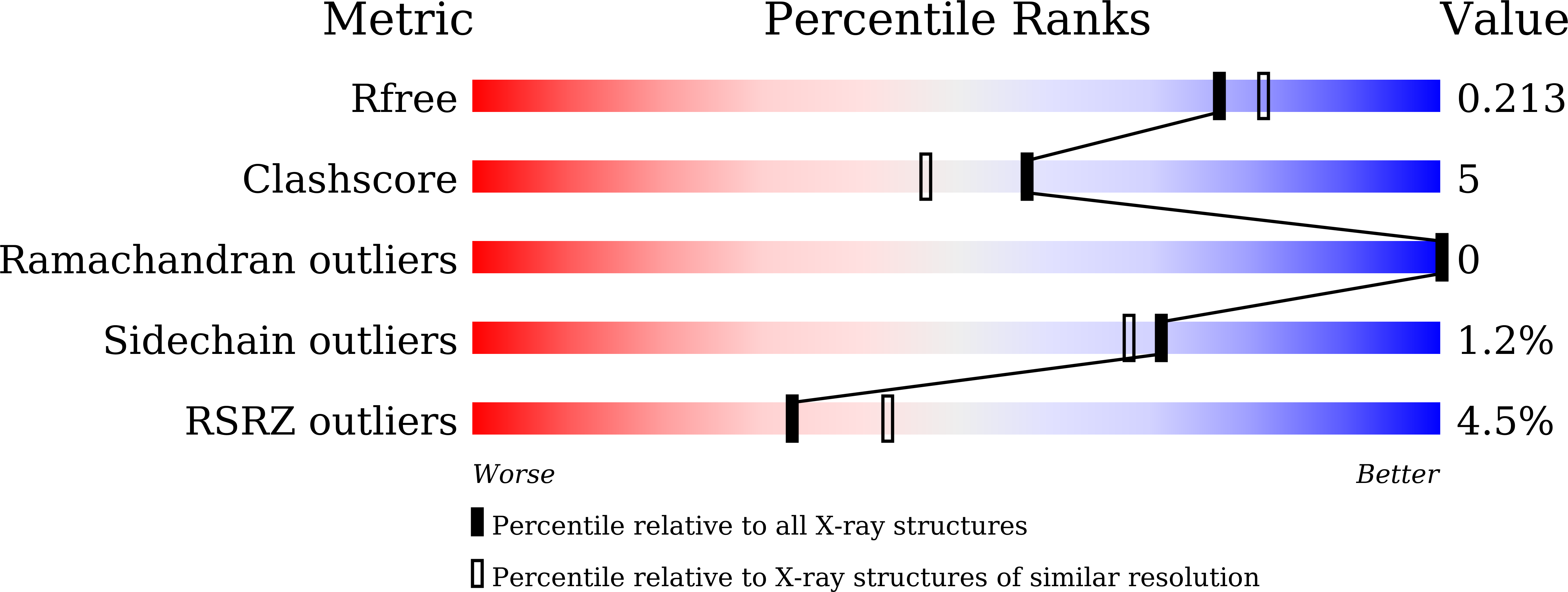
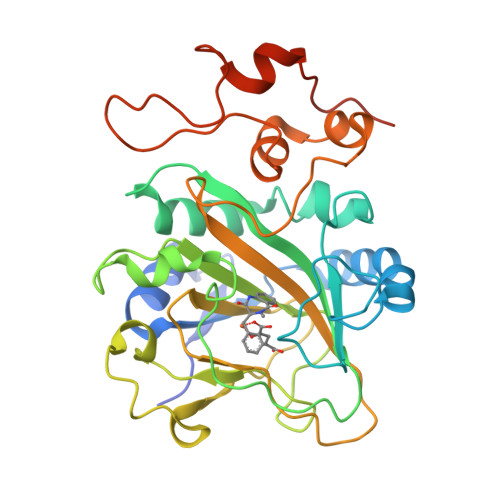
5Y7R, 5Y7T - PubMed Abstract:
AsqJ, an iron(II)- and 2-oxoglutarate-dependent enzyme found in viridicatin-type alkaloid biosynthetic pathways, catalyzes sequential desaturation and epoxidation to produce cyclopenins. Crystal structures of AsqJ bound to cyclopeptin and its C3 epimer are reported. Meanwhile, a detailed mechanistic study was carried out to decipher the desaturation mechanism. These findings suggest that a pathway involving hydrogen atom abstraction at the C10 position of the substrate by a short-lived Fe IV -oxo species and the subsequent formation of a carbocation or a hydroxylated intermediate is preferred during AsqJ-catalyzed desaturation.
Organizational Affiliation:
Institute of Biochemistry and Molecular Biology, College of Medicine, National (Taiwan) University, Taipei, 100, Taiwan.