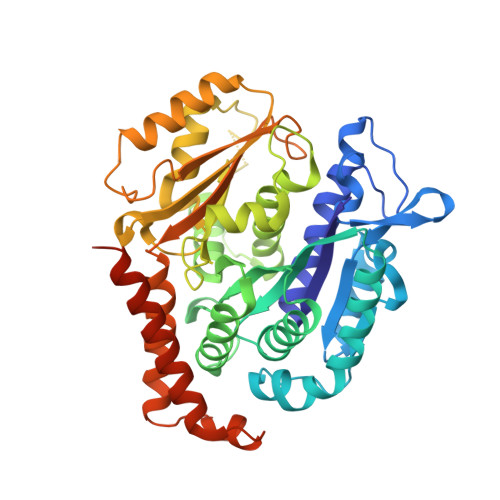
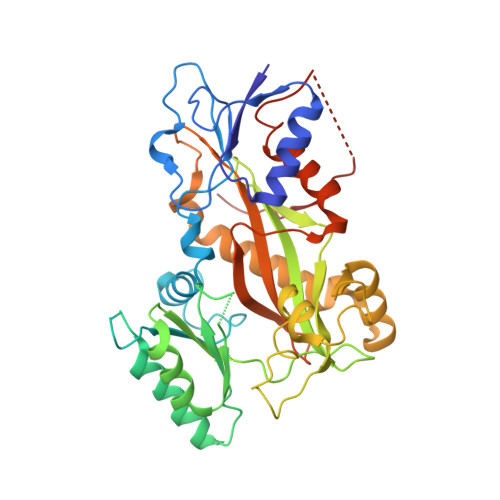
Comprehensive Analysis of Binding Sites in Tubulin.
Muhlethaler, T., Gioia, D., Prota, A.E., Sharpe, M.E., Cavalli, A., Steinmetz, M.O.(2021) Angew Chem Int Ed Engl 60: 13331-13342
- PubMed: 33951246
- DOI: https://doi.org/10.1002/anie.202100273
- Primary Citation of Related Structures:
5S4L, 5S4M, 5S4N, 5S4O, 5S4P, 5S4Q, 5S4R, 5S4S, 5S4T, 5S4U, 5S4V, 5S4W, 5S4X, 5S4Y, 5S4Z, 5S50, 5S51, 5S52, 5S53, 5S54, 5S55, 5S56, 5S57, 5S58, 5S59, 5S5A, 5S5B, 5S5C, 5S5D, 5S5E, 5S5F, 5S5G, 5S5H, 5S5I, 5S5J, 5S5K, 5S5L, 5S5M, 5S5N, 5S5O, 5S5P, 5S5Q, 5S5R, 5S5S, 5S5T, 5S5U, 5S5V, 5S5W, 5S5X, 5S5Y - PubMed Abstract:
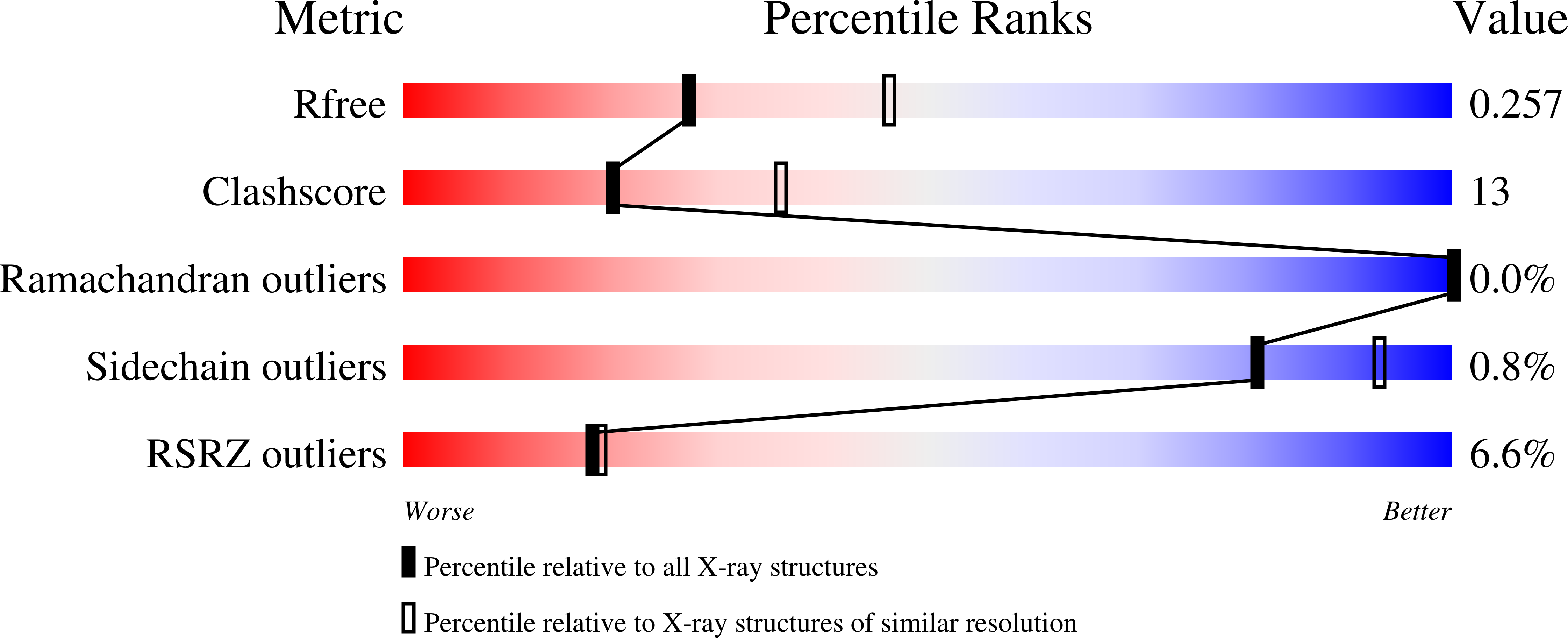
Tubulin plays essential roles in vital cellular activities and is the target of a wide range of proteins and ligands. Here, using a combined computational and crystallographic fragment screening approach, we addressed the question of how many binding sites exist in tubulin. We identified 27 distinct sites, of which 11 have not been described previously, and analyzed their relationship to known tubulin-protein and tubulin-ligand interactions. We further observed an intricate pocket communication network and identified 56 chemically diverse fragments that bound to 10 distinct tubulin sites. Our results offer a unique structural basis for the development of novel small molecules for use as tubulin modulators in basic research applications or as drugs. Furthermore, our method lays down a framework that may help to discover new pockets in other pharmaceutically important targets and characterize them in terms of chemical tractability and allosteric modulation.
Organizational Affiliation:
Laboratory of Biomolecular Research, Department of Biology and Chemistry, Paul Scherrer Institut, 5232, Villigen PSI, Switzerland.