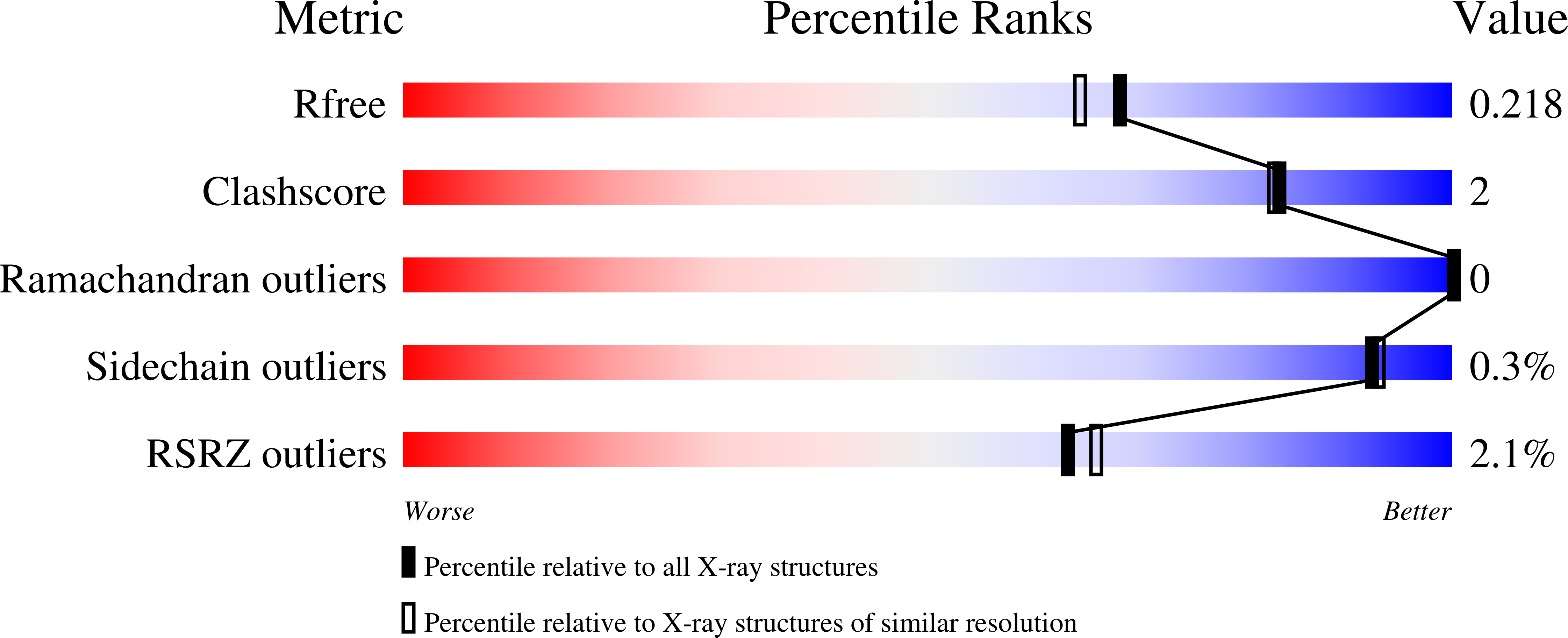
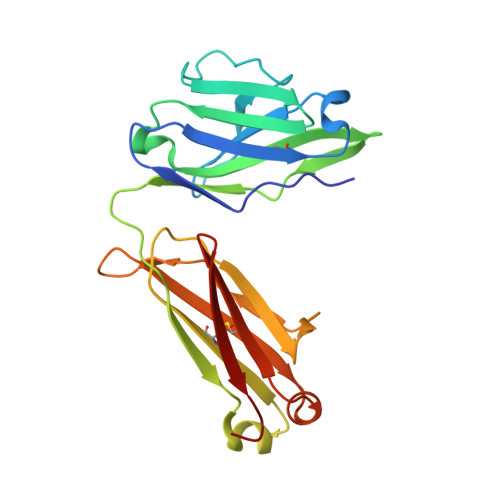
Rare PfCSP C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection.
Scally, S.W., Murugan, R., Bosch, A., Triller, G., Costa, G., Mordmuller, B., Kremsner, P.G., Sim, B.K.L., Hoffman, S.L., Levashina, E.A., Wardemann, H., Julien, J.P.(2018) J Exp Medicine 215: 63-75
- PubMed: 29167197
- DOI: https://doi.org/10.1084/jem.20170869
- Primary Citation of Related Structures:
6B0S, 6B0W - PubMed Abstract:
Antibodies against the central repeat of the Plasmodium falciparum (Pf) circumsporozoite protein (CSP) inhibit parasite activity and correlate with protection from malaria. However, the humoral response to the PfCSP C terminus (C-PfCSP) is less well characterized. Here, we describe B cell responses to C-PfCSP from European donors who underwent immunization with live Pf sporozoites (PfSPZ Challenge) under chloroquine prophylaxis (PfSPZ-CVac), and were protected against controlled human malaria infection. Out of 215 PfCSP-reactive monoclonal antibodies, only two unique antibodies were specific for C-PfCSP, highlighting the rare occurrence of C-PfCSP-reactive B cells in PfSPZ-CVac-induced protective immunity. These two antibodies showed poor sporozoite binding and weak inhibition of parasite traversal and development, and did not protect mice from infection with PfCSP transgenic Plasmodium berghei sporozoites. Structural analyses demonstrated that one antibody interacts with a polymorphic region overlapping two T cell epitopes, suggesting that variability in C-PfCSP may benefit parasite escape from humoral and cellular immunity. Our data identify important features underlying C-PfCSP shortcomings as a vaccine target.
Organizational Affiliation:
Program in Molecular Medicine, Hospital for Sick Children Research Institute, Toronto, Ontario, Canada.