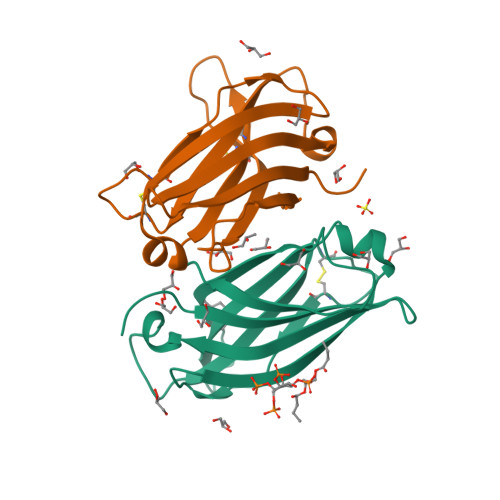
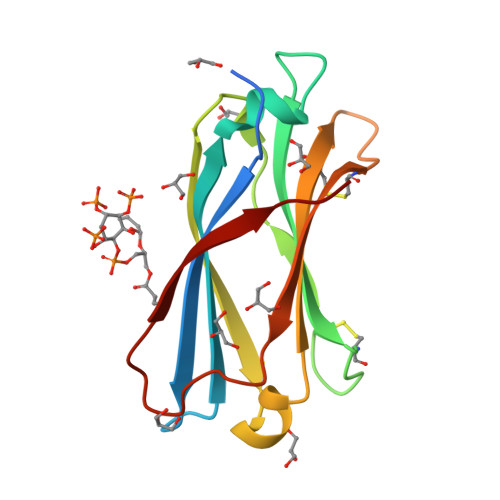
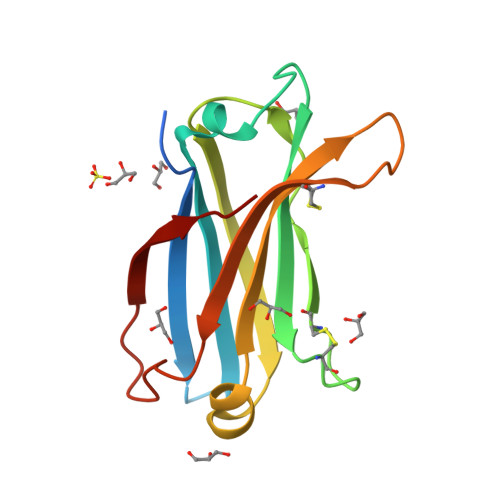
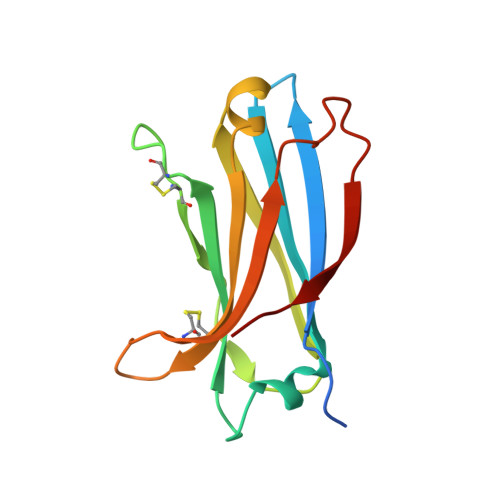
Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
Posner, M.G., Upadhyay, A., Ishima, R., Kalli, A.C., Harris, G., Kremerskothen, J., Sansom, M.S.P., Crennell, S.J., Bagby, S.(2018) J Biological Chem 293: 9335-9344
- PubMed: 29724824
- DOI: https://doi.org/10.1074/jbc.RA118.002279
- Primary Citation of Related Structures:
6FB4, 6FD0, 6FJC, 6FJD - PubMed Abstract:
Kidney- and brain-expressed protein (KIBRA), a multifunctional scaffold protein with around 20 known binding partners, is involved in memory and cognition, organ size control via the Hippo pathway, cell polarity, and membrane trafficking. KIBRA includes tandem N-terminal WW domains, a C2 domain, and motifs for binding atypical PKC and PDZ domains. A naturally occurring human KIBRA variant involving residue changes at positions 734 (Met-to-Ile) and 735 (Ser-to-Ala) within the C2 domain affects cognitive performance. We have elucidated 3D structures and calcium- and phosphoinositide-binding properties of human KIBRA C2 domain. Both WT and variant C2 adopt a canonical type I topology C2 domain fold. Neither Ca 2+ nor any other metal ion was bound to WT or variant KIBRA C2 in crystal structures, and Ca 2+ titration produced no significant reproducible changes in NMR spectra. NMR and X-ray diffraction data indicate that KIBRA C2 binds phosphoinositides via an atypical site involving β-strands 5, 2, 1, and 8. Molecular dynamics simulations indicate that KIBRA C2 interacts with membranes via primary and secondary sites on the same domain face as the experimentally identified phosphoinositide-binding site. Our results indicate that KIBRA C2 domain association with membranes is calcium-independent and involves distinctive C2 domain-membrane relative orientations.
Organizational Affiliation:
From the Department of Biology and Biochemistry, University of Bath, Bath BA2 7AY, United Kingdom.