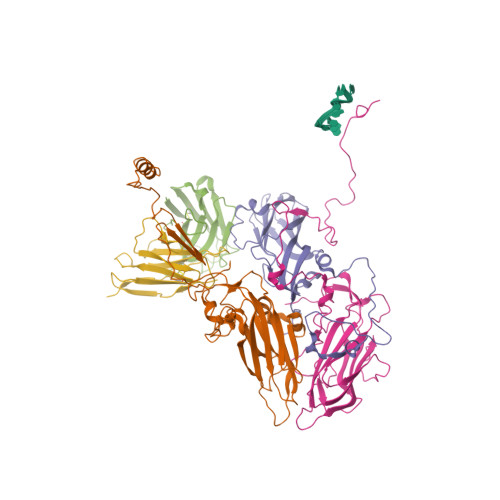
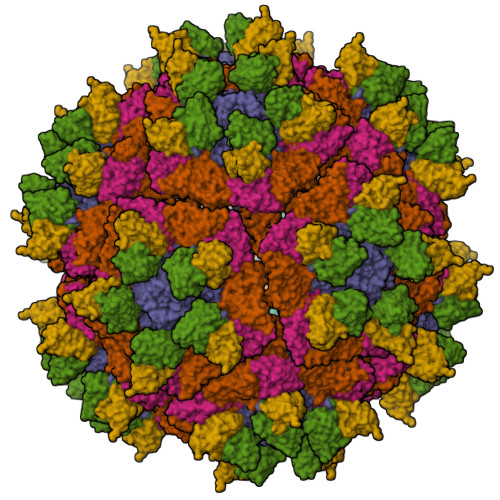
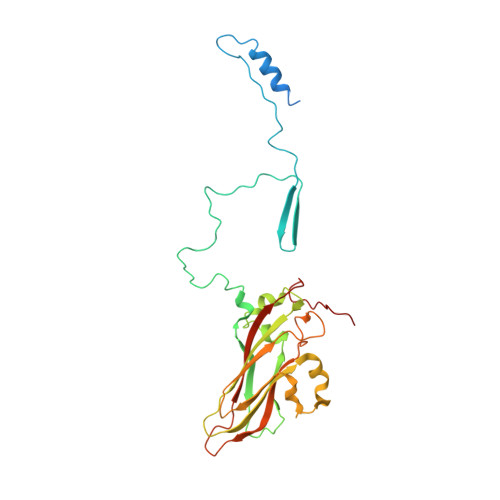
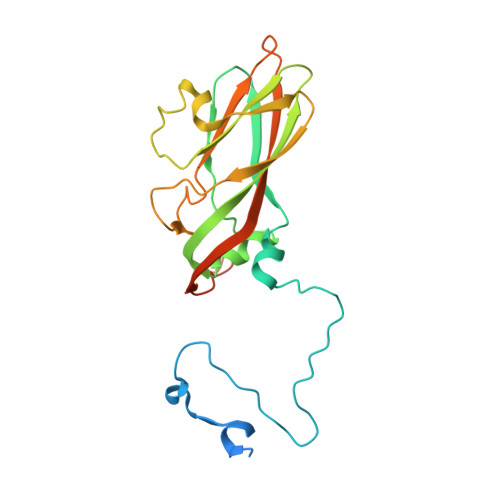
A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.
Domanska, A., Flatt, J.W., Jukonen, J.J.J., Geraets, J.A., Butcher, S.J.(2019) J Virol 93
- PubMed: 30463974
- DOI: https://doi.org/10.1128/JVI.01597-18
- Primary Citation of Related Structures:
6GV4 - PubMed Abstract:
Human parechovirus 3 (HPeV3) infection is associated with sepsis characterized by significant immune activation and subsequent tissue damage in neonates. Strategies to limit infection have been unsuccessful due to inadequate molecular diagnostic tools for early detection and the lack of a vaccine or specific antiviral therapy. Toward the latter, we present a 2.8-Å-resolution structure of HPeV3 in complex with fragments from a neutralizing human monoclonal antibody, AT12-015, using cryo-electron microscopy (cryo-EM) and image reconstruction. Modeling revealed that the epitope extends across neighboring asymmetric units with contributions from capsid proteins VP0, VP1, and VP3. Antibody decoration was found to block binding of HPeV3 to cultured cells. Additionally, at high resolution, it was possible to model a stretch of RNA inside the virion and, from this, identify the key features that drive and stabilize protein-RNA association during assembly. IMPORTANCE Human parechovirus 3 (HPeV3) is receiving increasing attention as a prevalent cause of sepsis-like symptoms in neonates, for which, despite the severity of disease, there are no effective treatments available. Structural and molecular insights into virus neutralization are urgently needed, especially as clinical cases are on the rise. Toward this goal, we present the first structure of HPeV3 in complex with fragments from a neutralizing monoclonal antibody. At high resolution, it was possible to precisely define the epitope that, when targeted, prevents virions from binding to cells. Such an atomic-level description is useful for understanding host-pathogen interactions and viral pathogenesis mechanisms and for finding potential cures for infection and disease.
Organizational Affiliation:
Faculty of Biological and Environmental Sciences, Molecular and Integrative Bioscience Research Programme, University of Helsinki, Helsinki, Finland ausra.domanska@helsinki.fi sarah.butcher@helsinki.fi.