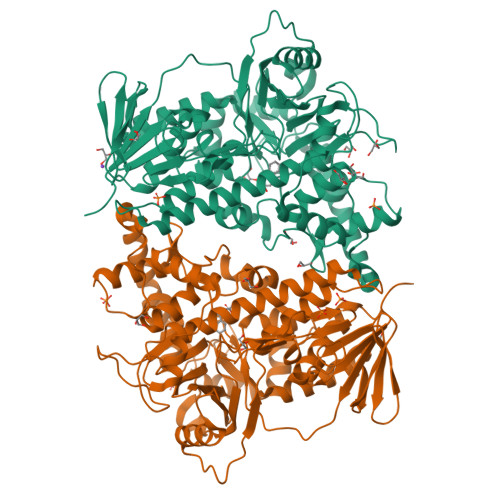
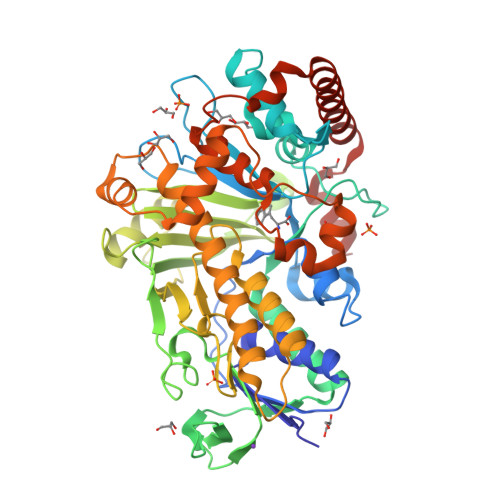
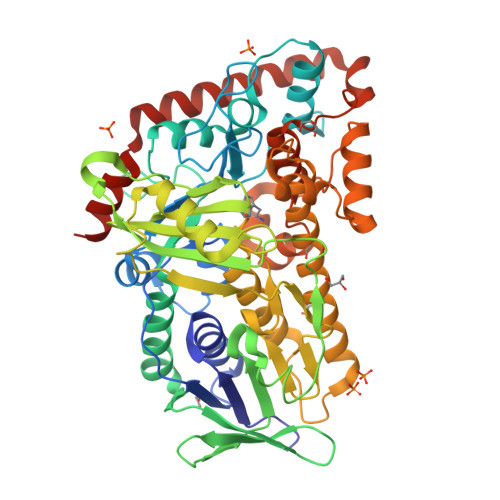
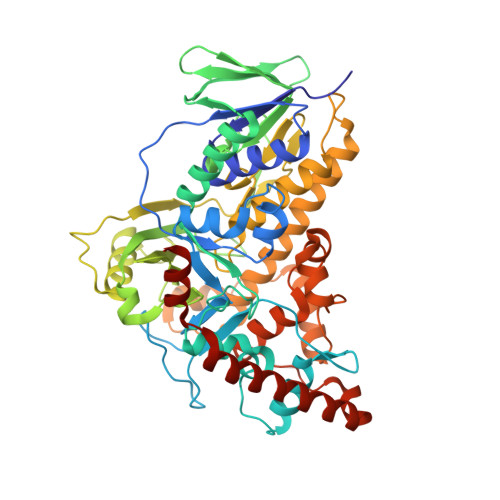
Structure-based switch of regioselectivity in the flavin-dependent tryptophan 6-halogenase Thal.
Moritzer, A.C., Minges, H., Prior, T., Frese, M., Sewald, N., Niemann, H.H.(2019) J Biological Chem 294: 2529-2542
- PubMed: 30559288
- DOI: https://doi.org/10.1074/jbc.RA118.005393
- Primary Citation of Related Structures:
6H43, 6H44, 6IB5 - PubMed Abstract:
Flavin-dependent halogenases increasingly attract attention as biocatalysts in organic synthesis, facilitating environmentally friendly halogenation strategies that require only FADH 2 , oxygen, and halide salts. Different flavin-dependent tryptophan halogenases regioselectively chlorinate or brominate trypto-phan's indole moiety at C5, C6, or C7. Here, we present the first substrate-bound structure of a tryptophan 6-halogenase, namely Thal, also known as ThdH, from the bacterium Streptomyces albogriseolus at 2.55 Å resolution. The structure revealed that the C6 of tryptophan is positioned next to the ϵ-amino group of a conserved lysine, confirming the hypothesis that proximity to the catalytic residue determines the site of electrophilic aromatic substitution. Although Thal is more similar in sequence and structure to the tryptophan 7-halogenase RebH than to the tryptophan 5-halogenase PyrH, the indole binding pose in the Thal active site more closely resembled that of PyrH than that of RebH. The difference in indole orientation between Thal and RebH appeared to be largely governed by residues positioning the Trp backbone atoms. The sequences of Thal and RebH lining the substrate binding site differ in only few residues. Therefore, we exchanged five amino acids in the Thal active site with the corresponding counterparts in RebH, generating the quintuple variant Thal-RebH5. Overall conversion of l-Trp by the Thal-RebH5 variant resembled that of WT Thal, but its regioselectivity of chlorination and bromination was almost completely switched from C6 to C7 as in RebH. We conclude that structure-based protein engineering with targeted substitution of a few residues is an efficient approach to tailoring flavin-dependent halogenases.
Organizational Affiliation:
From the Structural Biochemistry and.