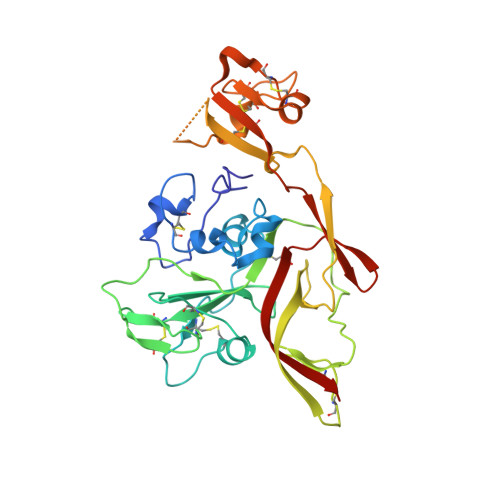
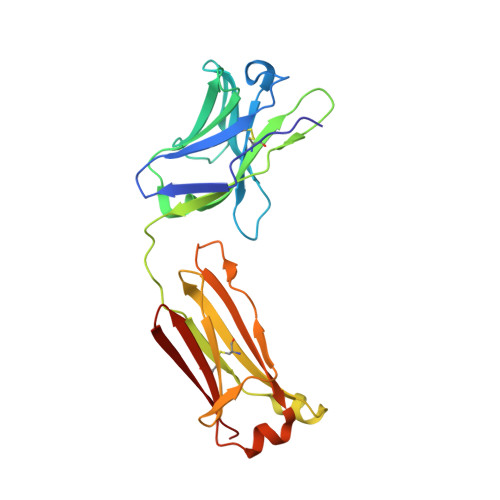
Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Wang, Q., Ma, T., Wu, Y., Chen, Z., Zeng, H., Tong, Z., Gao, F., Qi, J., Zhao, Z., Chai, Y., Yang, H., Wong, G., Bi, Y., Wu, L., Shi, R., Yang, M., Song, J., Jiang, H., An, Z., Wang, J., Yilma, T.D., Shi, Y., Liu, W.J., Liang, M., Qin, C., Gao, G.F., Yan, J.(2019) Nat Microbiol 4: 1231-1241
- PubMed: 30936489
- DOI: https://doi.org/10.1038/s41564-019-0411-z
- Primary Citation of Related Structures:
6IEA, 6IEB, 6IEC, 6IEK - PubMed Abstract:
Rift Valley fever virus (RVFV) is a mosquito-borne pathogen that causes substantial morbidity and mortality in livestock and humans. To date, there are no licensed human vaccines or therapeutics available. Here, we report the isolation of monoclonal antibodies from a convalescent patient, targeting the RVFV envelope proteins Gn and Gc. The Gn-specific monoclonal antibodies exhibited much higher neutralizing activities in vitro and protection efficacies in mice against RVFV infection, compared to the Gc-specific monoclonal antibodies. The Gn monoclonal antibodies were found to interfere with soluble Gn binding to cells and prevent infection by blocking the attachment of virions to host cells. Structural analysis of Gn complexed with four Gn-specific monoclonal antibodies resulted in the definition of three antigenic patches (A, B and C) on Gn domain I. Both patches A and B are major neutralizing epitopes. Our results highlight the potential of antibody-based therapeutics and provide a structure-based rationale for designing vaccines against RVFV.
Organizational Affiliation:
CAS Key Laboratory of Microbial Physiological and Metabolic Engineering, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.