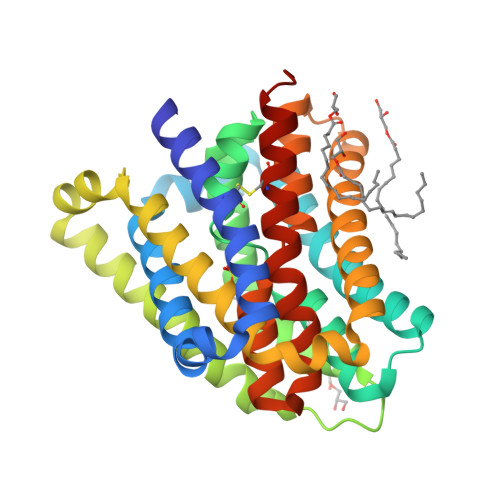
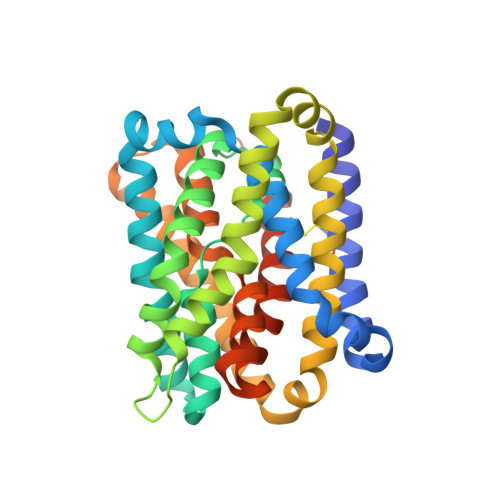
Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Wang, X., Lyu, Y., Ji, Y., Sun, Z., Zhou, X.(2021) Acta Crystallogr D Struct Biol 77: 117-125
- PubMed: 33404531
- DOI: https://doi.org/10.1107/S2059798320015004
- Primary Citation of Related Structures:
6LGV, 6LGY, 6LGZ, 6LH0 - PubMed Abstract:
Apical sodium-dependent bile acid transporter (ASBT) retrieves bile acids from the small intestine and plays a pivotal role in enterohepatic circulation. Currently, high-resolution structures are available for two bacterial ASBT homologs (ASBT NM from Neisseria meningitides and ASBT Yf from Yersinia frederiksenii), from which an elevator-style alternating-access mechanism has been proposed for substrate transport. A key concept in this model is that the substrate binds to the central cavity of the transporter so that the elevator-like motion can expose the bound substrate alternatingly to either side of the membrane during a transport cycle. However, no structure of an ASBT has been solved with a substrate bound in its central cavity, so how a substrate binds to ASBT remains to be defined. In this study, molecular docking, structure determination and functional analysis were combined to define and validate the details of substrate binding in ASBT Yf . The findings provide coherent evidence to provide a clearer picture of how the substrate binds in the central cavity of ASBT Yf that fits the alternating-access model.
Organizational Affiliation:
Department of Integrated Traditional Chinese and Western Medicine, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, People's Republic of China.