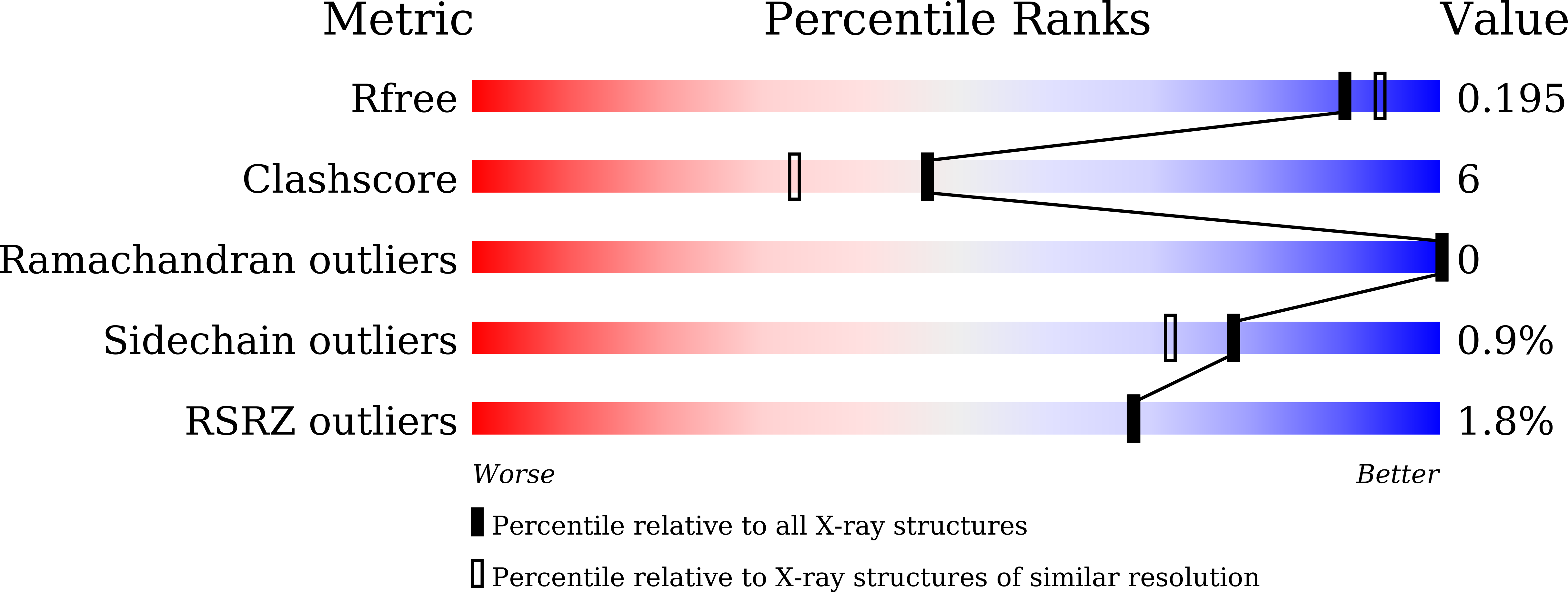
Structural basis for the broad substrate specificity of two acyl-CoA dehydrogenases FadE5 from mycobacteria.
Chen, X.B., Chen, J., Yan, B., Zhang, W., Guddat, L.W., Liu, X., Rao, Z.(2020) Proc Natl Acad Sci U S A 117: 16324-16332
- PubMed: 32601219
- DOI: https://doi.org/10.1073/pnas.2002835117
- Primary Citation of Related Structures:
6KPT, 6KRI, 6KS9, 6KSA, 6KSB, 6KSE, 6LPY, 6LQ0, 6LQ1, 6LQ2, 6LQ3, 6LQ4, 6LQ5, 6LQ6, 6LQ7, 6LQ8 - PubMed Abstract:
FadE, an acyl-CoA dehydrogenase, introduces unsaturation to carbon chains in lipid metabolism pathways. Here, we report that FadE5 from Mycobacterium tuberculosis ( Mtb FadE5) and Mycobacterium smegmatis ( Ms FadE5) play roles in drug resistance and exhibit broad specificity for linear acyl-CoA substrates but have a preference for those with long carbon chains. Here, the structures of Ms FadE5 and Mtb FadE5, in the presence and absence of substrates, have been determined. These reveal the molecular basis for the broad substrate specificity of these enzymes. FadE5 interacts with the CoA region of the substrate through a large number of hydrogen bonds and an unusual π-π stacking interaction, allowing these enzymes to accept both short- and long-chain substrates. Residues in the substrate binding cavity reorient their side chains to accommodate substrates of various lengths. Longer carbon-chain substrates make more numerous hydrophobic interactions with the enzyme compared with the shorter-chain substrates, resulting in a preference for this type of substrate.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, Frontiers Science Center for Cell Responses, College of Life Sciences, Nankai University, 300071 Tianjin, China.