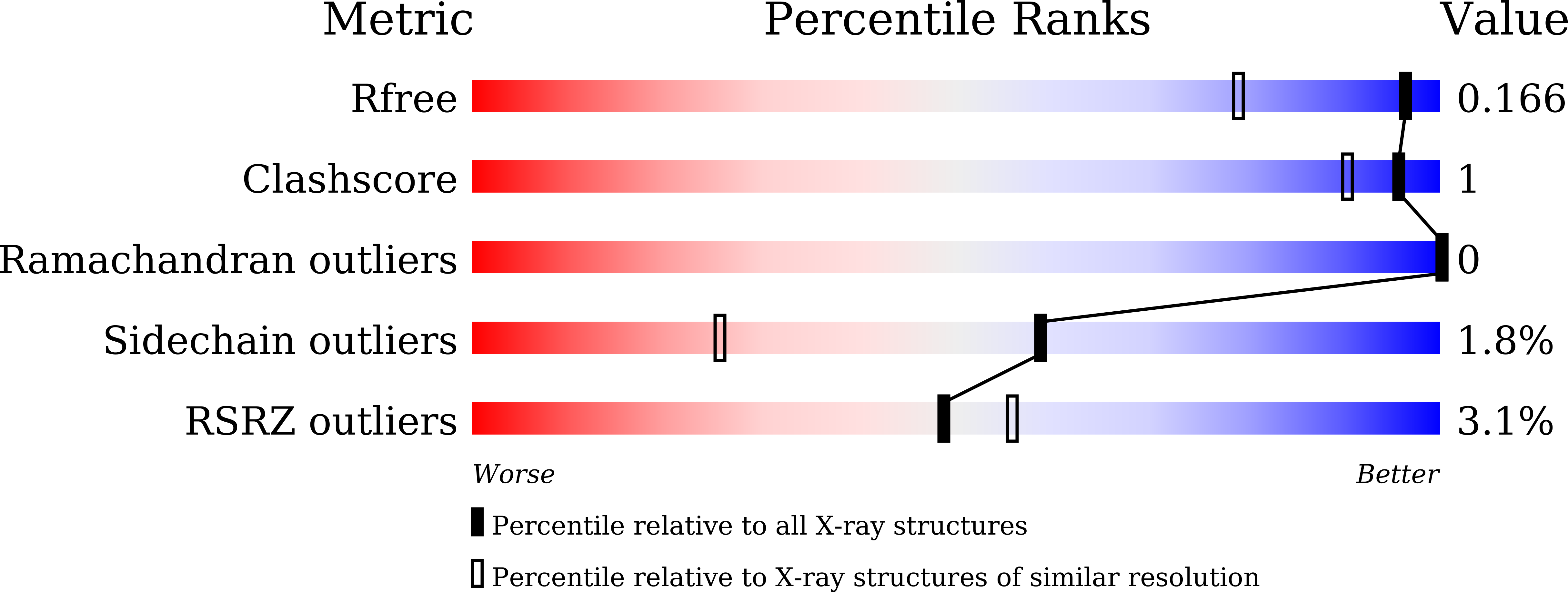
Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Tonozuka, T., Kitamura, J., Nagaya, M., Kawai, R., Nishikawa, A., Hirano, K., Tamura, K., Fujii, T., Tochio, T.(2020) Biosci Biotechnol Biochem 84: 2508-2520
- PubMed: 32752982
- DOI: https://doi.org/10.1080/09168451.2020.1804317
- Primary Citation of Related Structures:
6M0D, 6M0E - PubMed Abstract:
An enzyme belonging to glycoside hydrolase family 68 (GH68) from Beijerinckia indica subsp. indica NBRC 3744 was expressed in Escherichia coli . Biochemical characterization showed that the enzyme was identified to be a β-fructosyltransferase (BiBftA). Crystallization of a full-length BiBftA was initially attempted, but no crystals were obtained. We constructed a variant in which 5 residues (Pro199-Gly203) and 13 residues (Leu522-Gln534) in potentially flexible regions were deleted, and we successfully crystallized this variant BiBftA. BiBftA is composed of a five-bladed β-propeller fold as in other GH68 enzymes. The structure of BiBftA in complex with fructose unexpectedly indicated that one β-fructofuranose (β-Fru f ) molecule and one β-fructopyranose molecule bind to the catalytic pocket. The orientation of β-Fru f at subsite -1 is tilted from the orientation observed in most GH68 enzymes, presenting a second structure of a GH68 enzyme in complex with the tilted binding mode of β-Fru f.
Organizational Affiliation:
Department of Applied Biological Science, Tokyo University of Agriculture and Technology , Tokyo, Japan.