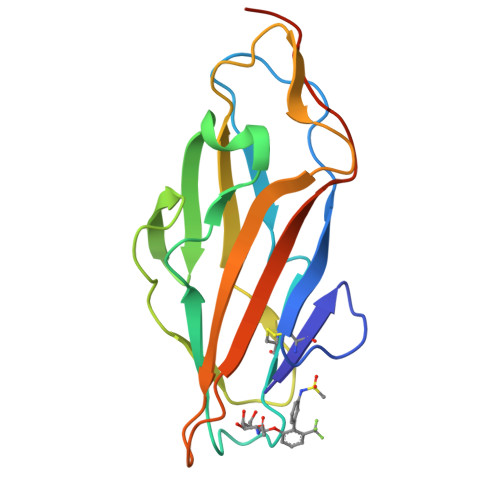
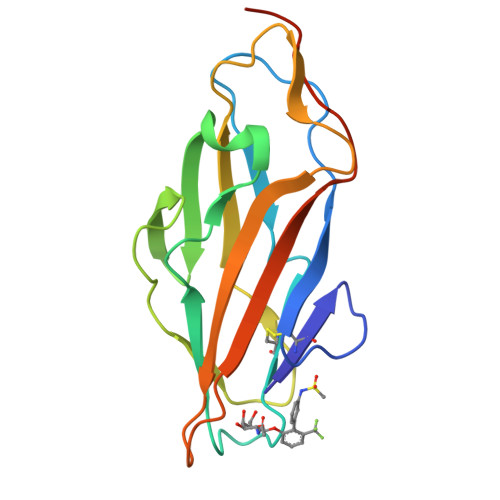
Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
Maddirala, A.R., Klein, R., Pinkner, J.S., Kalas, V., Hultgren, S.J., Janetka, J.W.(2019) J Med Chem 62: 467-479
- PubMed: 30540910
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01561
- Primary Citation of Related Structures:
6MAP, 6MAQ, 6MAW - PubMed Abstract:
The F9/Yde/Fml pilus, tipped with the FmlH adhesin, has been shown to provide uropathogenic Escherichia coli (UPEC) a fitness advantage in urinary tract infections (UTIs). Here, we used X-ray structure guided design to optimize our previously described ortho-biphenyl Gal and GalNAc FmlH antagonists such as compound 1 by replacing the carboxylate with a sulfonamide as in 50. Other groups which can accept H-bonds were also tolerated. We pursued further modifications to the biphenyl aglycone resulting in significantly improved activity. Two of the most potent compounds, 86 (IC 50 = 0.051 μM) and 90 (IC 50 = 0.034 μM), exhibited excellent metabolic stability in mouse plasma and liver microsomes but showed only limited oral bioavailability (<1%) in rats. Compound 84 also showed a good pharmacokinetic (PK) profile in mice after IP dosing with compound exposure above the IC 50 for 6 h. These new FmlH antagonists represent new antivirulence drugs for UTIs.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics , Washington University School of Medicine , St. Louis , Missouri 63110 , United States.