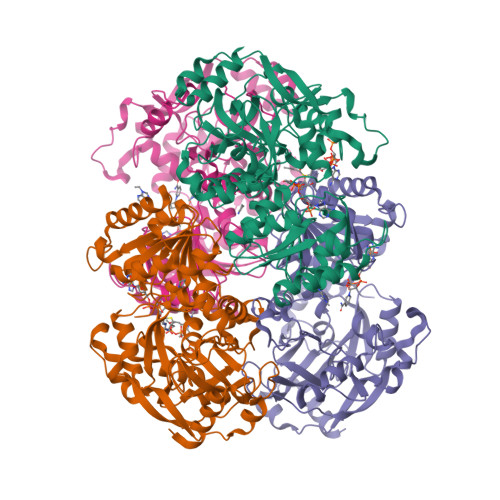
Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
DeMirci, H., Rao, Y., Stoffel, G.M., Vogeli, B., Schell, K., Gomez, A., Batyuk, A., Gati, C., Sierra, R.G., Hunter, M.S., Dao, E.H., Ciftci, H.I., Hayes, B., Poitevin, F., Li, P.N., Kaur, M., Tono, K., Saez, D.A., Deutsch, S., Yoshikuni, Y., Grubmuller, H., Erb, T.J., Vohringer-Martinez, E., Wakatsuki, S.(2022) ACS Cent Sci