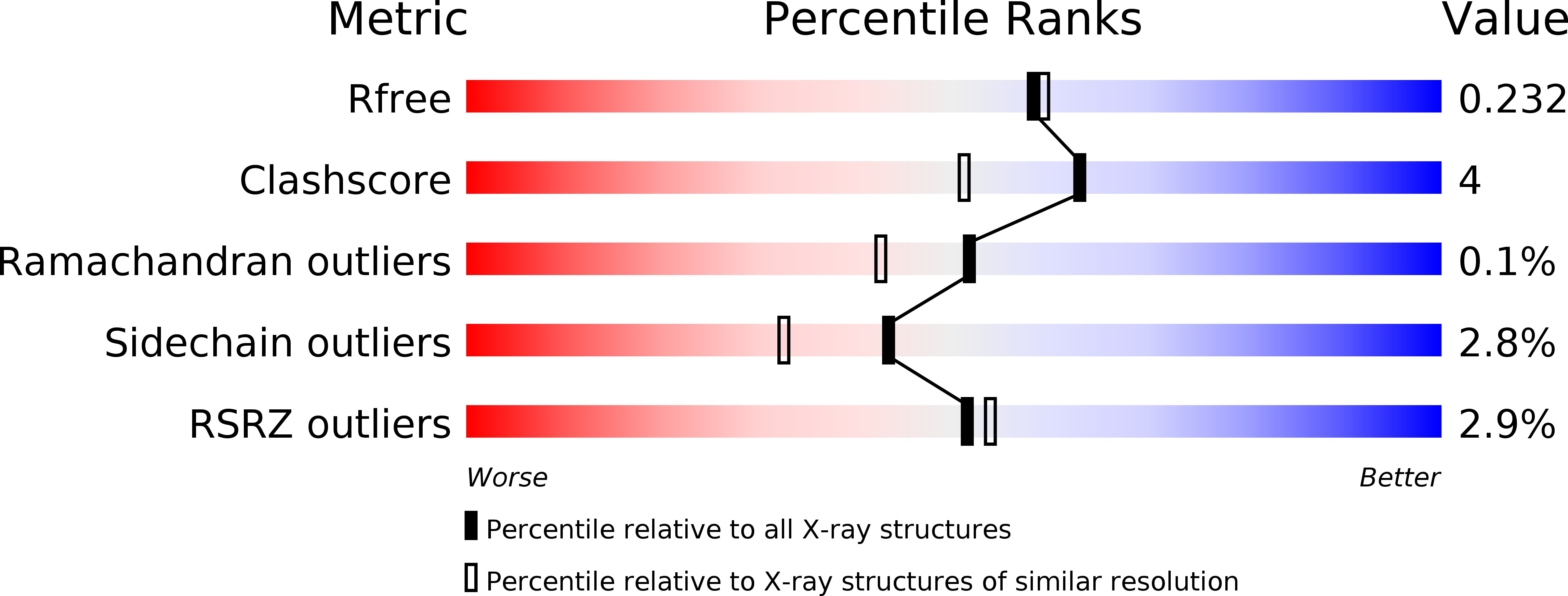
Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Lubkowski, J., Chan, W., Wlodawer, A.(2019) Sci Rep 9: 11070-11070
- PubMed: 31363102
- DOI: https://doi.org/10.1038/s41598-019-46432-0
- Primary Citation of Related Structures:
6NX6, 6NX7, 6NX8, 6NX9, 6NXA, 6NXB, 6NXC, 6NXD - PubMed Abstract:
Active sites of enzymes are highly optimized for interactions with specific substrates, thus binding of opportunistic ligands is usually observed only in the absence of native substrates or products. However, during growth of crystals required for structure determination enzymes are often exposed to conditions significantly divergent from the native ones, leading to binding of unexpected ligands to active sites even in the presence of substrates. Failing to recognize this possibility may lead to incorrect interpretation of experimental results and to faulty conclusions. Here, we present several examples of binding of a citrate anion to the active sites of E. coli L-asparaginases I and II, even in the presence of the native substrate, L-Asn. A part of this report focuses on a comprehensive re-interpretation of structural results published previously for complexes of type I L-asparaginase (EcAI) from E. coli. In two re-refined structures a citrate anion forms an acyl-enzyme reaction intermediate with the catalytic threonine. These results emphasize the importance of careful and critical analysis during interpretation of crystallographic data.
Organizational Affiliation:
Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, 21702, USA. lubkowsj@mail.nih.gov.