GLMP is essential for bone-marrow hematopoiesis and lysosomal glycolipid metabolism
Manzanillo, P., Huang, C.S., Boenig, G., Calses, P., Scherl, A., Reichelt, M., Katakam, A.K., Martin, F., Hymowitz, S.G., Ouyang, W.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 1H3 Fab light chain | A [auth L] | 214 | Mus musculus | Mutation(s): 0 | 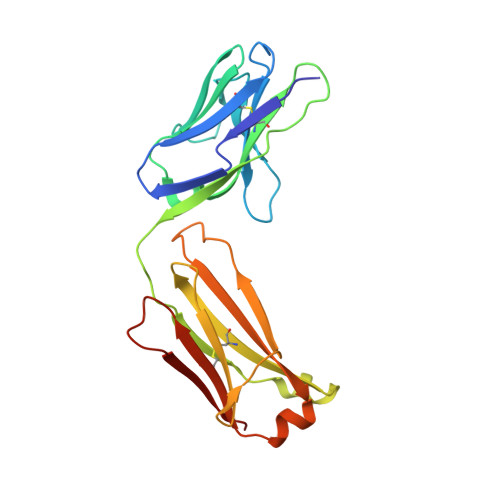 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 1H3 Fab heavy chain | B [auth H] | 227 | Mus musculus | Mutation(s): 0 | 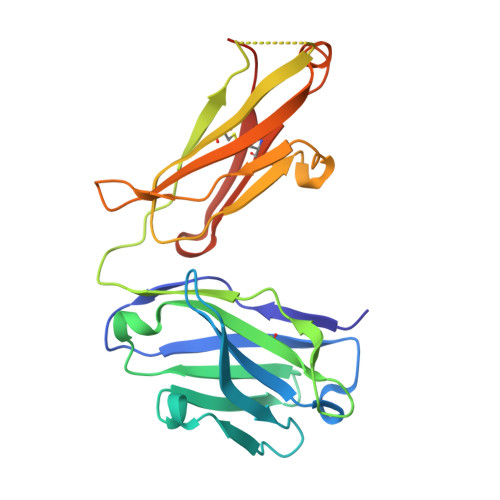 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycosylated lysosomal membrane protein | 404 | Mus musculus | Mutation(s): 0 Gene Names: Glmp | 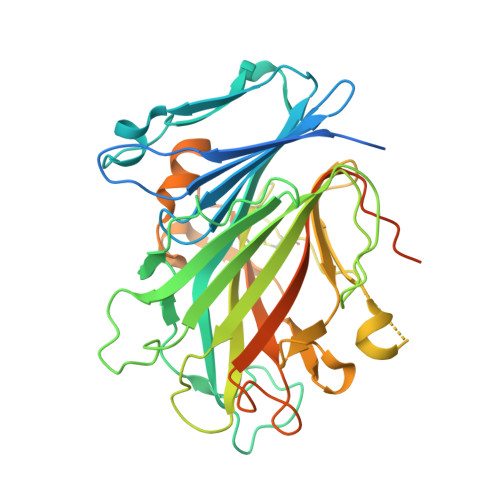 | |
UniProt | |||||
Find proteins for Q9JHJ3 (Mus musculus) Explore Q9JHJ3 Go to UniProtKB: Q9JHJ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9JHJ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | H [auth C] I [auth C] J [auth C] K [auth C] L [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| NA Query on NA | D [auth H], E [auth C], F [auth C], G [auth C] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.924 | α = 90 |
| b = 55.238 | β = 99.35 |
| c = 99.639 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |