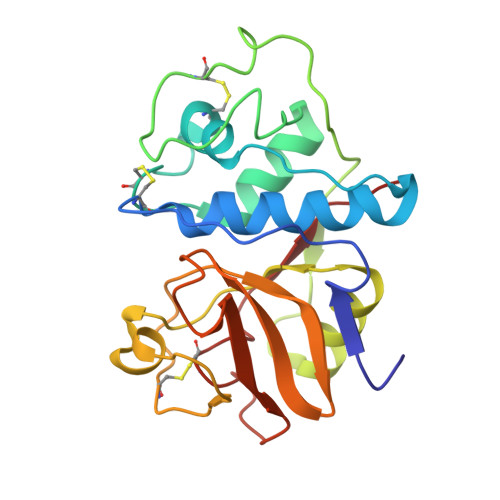
Binding of chloromethyl ketone substrate analogues to crystalline papain.
Drenth, J., Kalk, K.H., Swen, H.M.(1976) Biochemistry 15: 3731-3738
- PubMed: 952885
- DOI: https://doi.org/10.1021/bi00662a014
- Primary Citation of Related Structures:
1PAD, 2PAD, 4PAD, 5PAD, 6PAD - PubMed Abstract:
Papain (EC 3.4.22.2) is a proteolytic enzyme, the three-dimensional structure of which has been determined by x-ray diffraction at 2.8 A resolution (Drenth, J., Jansonius, J.N., Koekoek, R., Swen, H. M., and Wothers, B.G. (1968), Nature (London) 218, 929-932). The active site is a groove on the molecular surface in which the essential sulfhydryl group of cysteine-25 is situated next to the imidazole ring of histidine-159. The main object of this study was to determine by the difference-Fourier technique the binding mode for the substrate in the groove in order to explain the substrate specificity of the enzyme (P2 should have a hydrophobic side chain (Berger and Schechter, 1970) and to contribute to an elucidation of the catalytic mechanism. To this end, three chloromethyl ketone substrate analogues were reacted with the enzyme by covalent attachment to the sulfur atom of cysteine-25. The products crystallized isomorphously with the parent structure that is not the native, active enzyme but a mixture of oxidized papain (probably papain-SO2-) and papain with an extra cysteine attached to cysteine-25. Although this made the interpretation of the difference electron density maps less easy, it provided us with a clear picture of the way in which the acyl part of the substrate binds in the active site groove. The carbonyl oxygen of the P1 residue is near two potential hydrogen-bond donating groups, the backbone NH of cysteine-25 and the NH2 of glutamine-19. Valine residues 133 and 157 are responsible for the preference of papain in its substrate splitting. By removing the methylene group that covalently attaches the inhibitor molecules to the sulfur atom of cysteine-25 we obtained acceptable models for the acyl-enzyme structure and for the tetrahedral intermediate. The carbonyl oxygen of the P1 residue, carrying a formal negative charge in the tetrahedral intermediate, is stabilized by formation of two hydrogen bonds with the backbone NH of cysteine-25 and the NH2 group of glutamine-19. This situation resembles that suggested for the proteolytic serine enzymes (Henderson, R., Wright, C. S., Hess, G. P., and Blow, D. M. (1971), Cold Spring Harbor Symp. Quant. Biol. 36, 63-70; Robertus, J. D., Kraut, J., Alden, R. A., and Birktoft, J. J. (1972b), Biochemistry 11, 4293-4303). The nitrogen atom of the scissile peptide bond was found close to the imidazole ring of histidine-159, suggesting a role for this ring in protonating the N atom of the leaving group (Lowe, 1970). This proton transfer would be facilitated by a 30 degrees rotation of the ring around the C beta-Cgamma bond from an in-plane position with the sulfur atom to an in-plane position with the N atom. The possibility of this rotation is derived from a difference electron-density map for fully oxidizied papain vs. the parent protein.