Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Gassler, C.S., Ryan, M., Liu, T., Griffith, O.H., Heinz, D.W.(1997) Biochemistry 36: 12802-12813
- PubMed: 9335537
- DOI: https://doi.org/10.1021/bi971102d
- Primary Citation of Related Structures:
2PTD, 3PTD, 4PTD, 5PTD, 6PTD, 7PTD - PubMed Abstract:
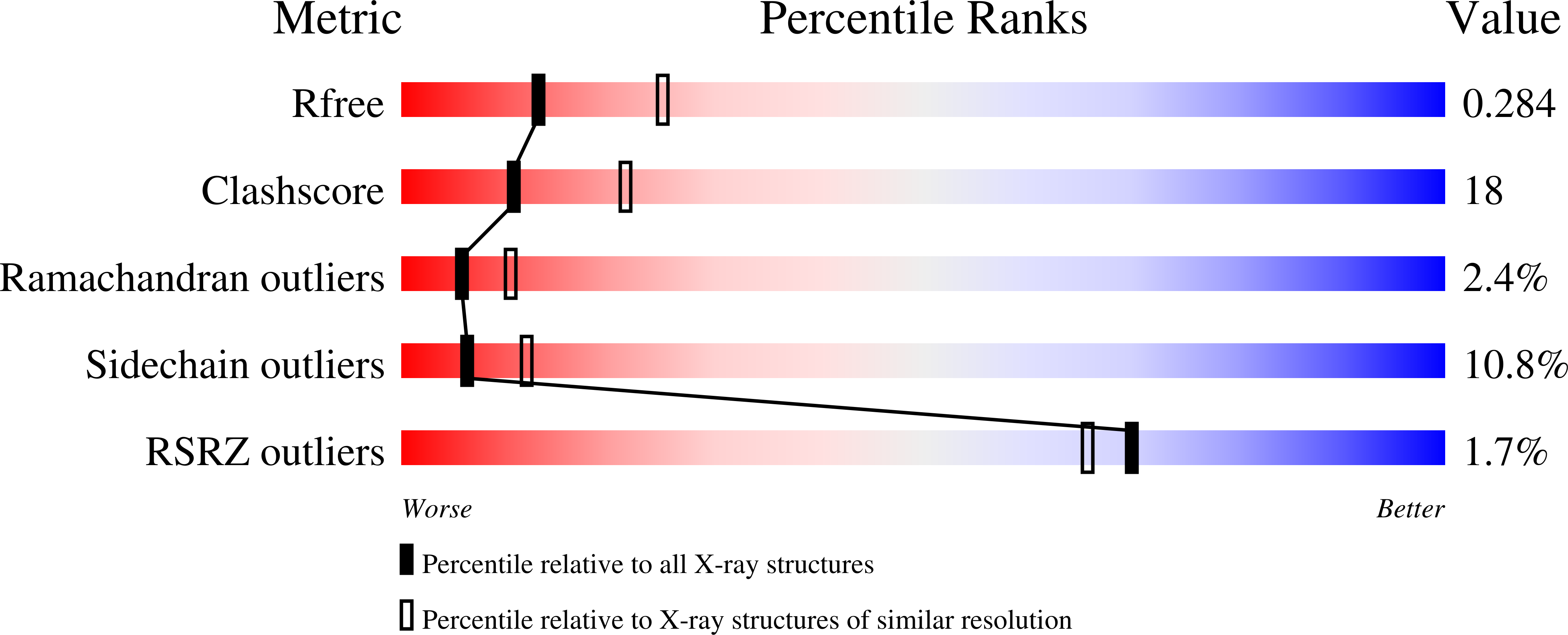
The role of amino acid residues located in the active site pocket of phosphatidylinositol-specific phospholipase C (PI-PLC) from Bacillus cereus[Heinz, D. W., Ryan, M., Bullock, T., & Griffith, O. H. (1995) EMBO J. 14, 3855-3863] was investigated by site-directed mutagenesis, kinetics, and crystal structure analysis. Twelve residues involved in catalysis and substrate binding (His32, Arg69, His82, Gly83, Lys115, Glu117, Arg163, Trp178, Asp180, Asp198, Tyr200, and Asp274) were individually replaced by 1-3 other amino acids, resulting in a total number of 21 mutants. Replacements in the mutants H32A, H32L, R69A, R69E, R69K, H82A, H82L, E117K, R163I, D198A, D198E, D198S, Y200S, and D274S caused essentially complete inactivation of the enzyme. The remaining mutants (G83S, K115E, R163K, W178Y, D180S, Y200F, and D274N) exhibited reduced activities up to 57% when compared with wild-type PI-PLC. Crystal structures determined at a resolution ranging from 2.0 to 2.7 A for six mutants (H32A, H32L, R163K, D198E, D274N, and D274S) showed that significant changes were confined to the site of the respective mutation without perturbation of the rest of the structure. Only in mutant D198E do the side chains of two neighboring arginine residues move across the inositol binding pocket toward the newly introduced glutamic acid. An analysis of these structure-function relationships provides new insight into the catalytic mechanism, and suggests a molecular explanation of some of the substrate stereospecificity and inhibitor binding data available for this enzyme.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Universität Freiburg, D-79104 Freiburg, Germany.