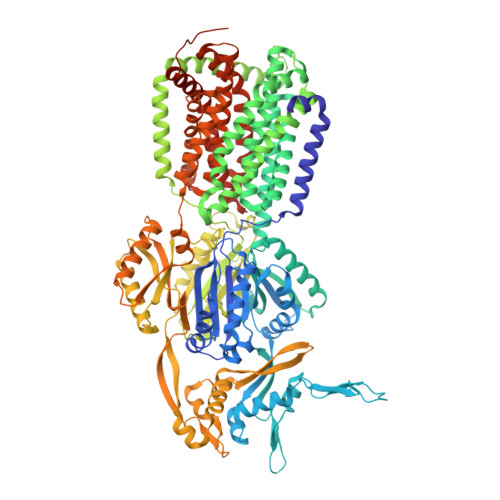
Binding and Transport of Carboxylated Drugs by the Multidrug Transporter AcrB.
Tam, H.K., Malviya, V.N., Foong, W.E., Herrmann, A., Malloci, G., Ruggerone, P., Vargiu, A.V., Pos, K.M.(2020) J Mol Biol 432: 861-877
- PubMed: 31881208
- DOI: https://doi.org/10.1016/j.jmb.2019.12.025
- Primary Citation of Related Structures:
6Q4N, 6Q4O, 6Q4P - PubMed Abstract:
AcrAB(Z)-TolC is the main drug efflux transporter complex in Escherichia coli. The extrusion of various toxic compounds depends on several drug binding sites within the trimeric AcrB transporter. Membrane-localized carboxylated substrates, such as fusidic acid and hydrophobic β-lactams, access the pump via a groove between the transmembrane helices TM1 and TM2. In this article, the transport route from the initial TM1/TM2 groove binding site toward the deep binding pocket located in the periplasmic part has been addressed via molecular modeling studies followed by functional and structural characterization of several AcrB variants. We propose that membrane-embedded drugs bind initially to the TM1/TM2 groove, are oriented by the AcrB PN2 subdomain, and are subsequently transported via a PN2/PC1 interface pathway directly toward the deep binding pocket. Our work emphasizes the exploitation of multiple transport pathways by AcrB tuned to substrate physicochemical properties related to the polyspecificity of the pump.
Organizational Affiliation:
Institute of Biochemistry, Goethe University Frankfurt, Max-von-Laue-Str. 9, D-60438 Frankfurt Am Main, Germany.