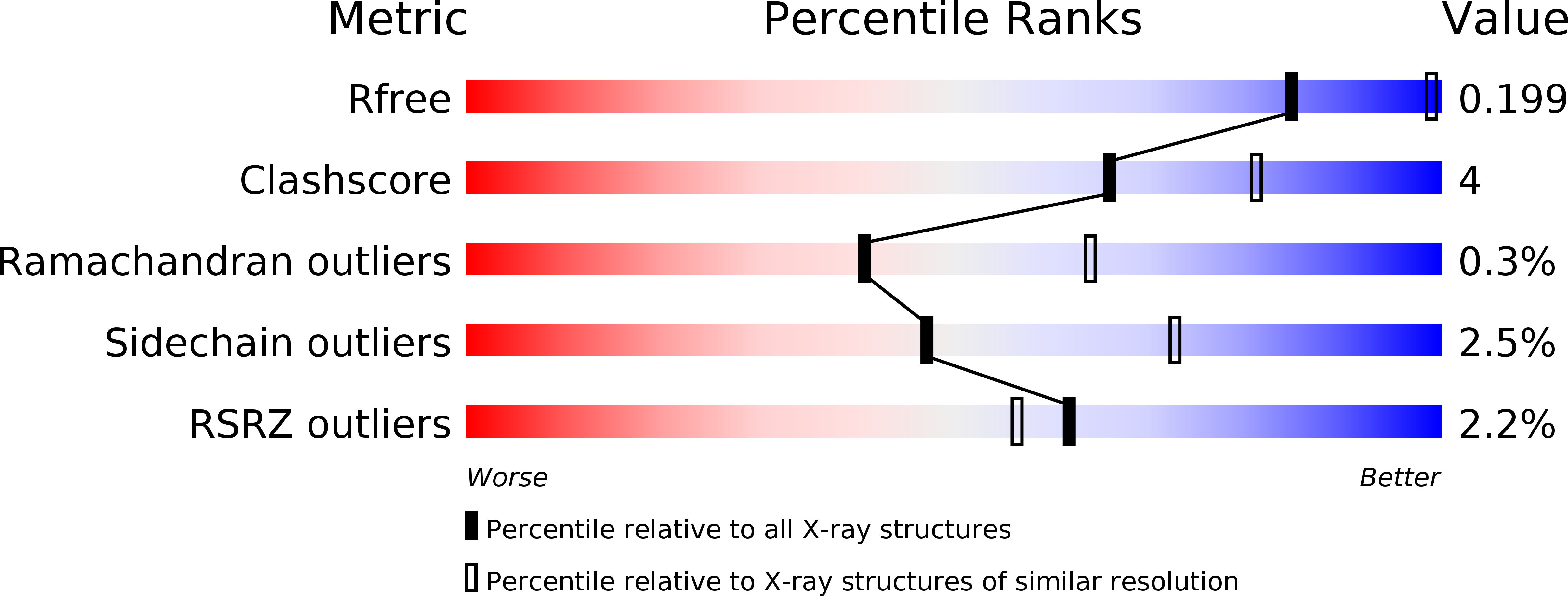
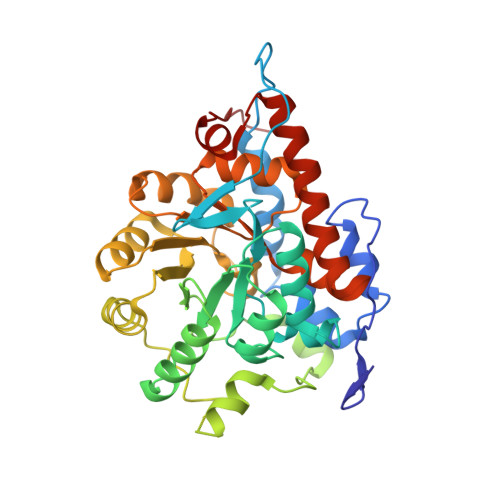
FMN-dependent oligomerization of putative lactate oxidase from Pediococcus acidilactici.
Ashok, Y., Maksimainen, M.M., Kallio, T., Kilpelainen, P., Lehtio, L.(2020) PLoS One 15: e0223870-e0223870
- PubMed: 32092083
- DOI: https://doi.org/10.1371/journal.pone.0223870
- Primary Citation of Related Structures:
6R9V, 6RHS, 6RHT - PubMed Abstract:
Lactate oxidases belong to a group of FMN-dependent enzymes and they catalyze a conversion of lactate to pyruvate with a release of hydrogen peroxide. Hydrogen peroxide is also utilized as a read out in biosensors to quantitate lactate levels in biological samples. Aerococcus viridans lactate oxidase is the best characterized lactate oxidase and our knowledge of lactate oxidases relies largely to studies conducted with that particular enzyme. Pediococcus acidilactici lactate oxidase is also commercially available for e.g. lactate measurements, but this enzyme has not been characterized in detail before. Here we report structural characterization of the recombinant enzyme and its co-factor dependent oligomerization. The crystal structures revealed two distinct conformations in the loop closing the active site, consistent with previous biochemical studies implicating the role of loop in catalysis. Despite the structural conservation of active site residues, we were not able to detect either oxidase or monooxygenase activity when L-lactate was used as a substrate. Pediococcus acidilactici lactate oxidase is therefore an example of a misannotation of an FMN-dependent enzyme, which catalyzes likely a so far unknown oxidation reaction.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine, University of Oulu, Oulu, Finland.