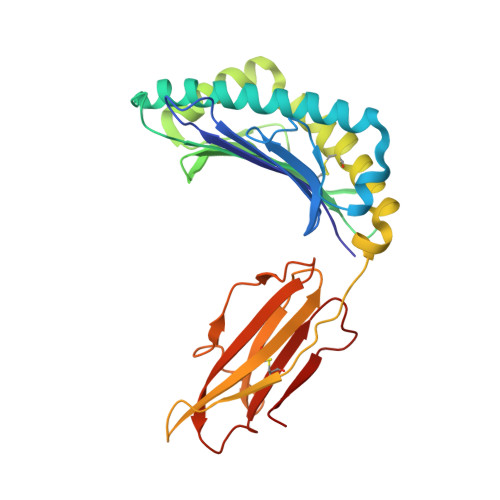
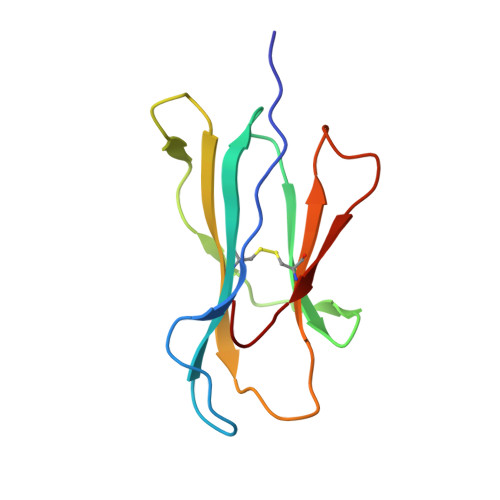
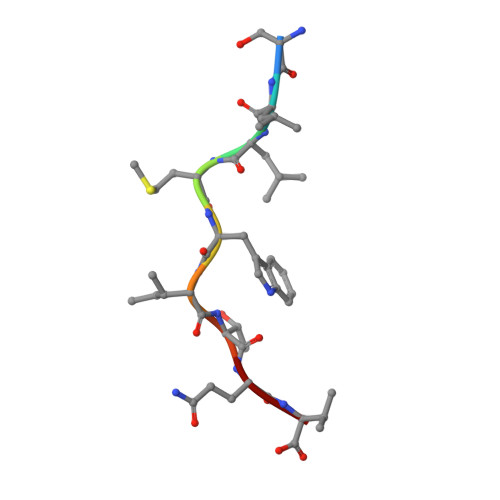
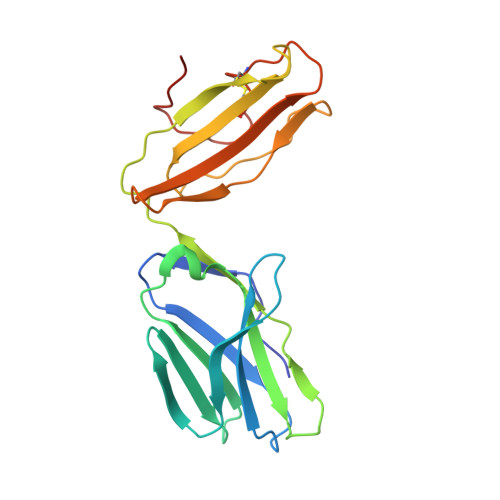
TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
Coles, C.H., Mulvaney, R.M., Malla, S., Walker, A., Smith, K.J., Lloyd, A., Lowe, K.L., McCully, M.L., Martinez Hague, R., Aleksic, M., Harper, J., Paston, S.J., Donnellan, Z., Chester, F., Wiederhold, K., Robinson, R.A., Knox, A., Stacey, A.R., Dukes, J., Baston, E., Griffin, S., Jakobsen, B.K., Vuidepot, A., Harper, S.(2020) J Immunol 204: 1943-1953
- PubMed: 32102902
- DOI: https://doi.org/10.4049/jimmunol.1900915
- Primary Citation of Related Structures:
6RP9, 6RPA, 6RPB - PubMed Abstract:
The molecular rules driving TCR cross-reactivity are poorly understood and, consequently, it is unclear the extent to which TCRs targeting the same Ag recognize the same off-target peptides. We determined TCR-peptide-HLA crystal structures and, using a single-chain peptide-HLA phage library, we generated peptide specificity profiles for three newly identified human TCRs specific for the cancer testis Ag NY-ESO-1 157-165 -HLA-A2. Two TCRs engaged the same central peptide feature, although were more permissive at peripheral peptide positions and, accordingly, possessed partially overlapping peptide specificity profiles. The third TCR engaged a flipped peptide conformation, leading to the recognition of off-target peptides sharing little similarity with the cognate peptide. These data show that TCRs specific for a cognate peptide recognize discrete peptide repertoires and reconciles how an individual's limited TCR repertoire following negative selection in the thymus is able to recognize a vastly larger antigenic pool.
Organizational Affiliation:
Immunocore, Ltd., Abingdon, Oxfordshire OX14 4RY, United Kingdom; and.