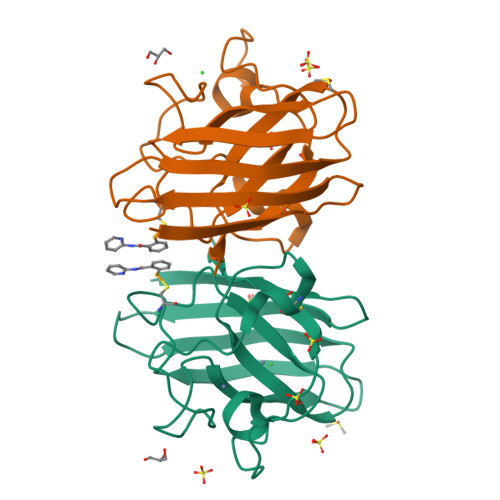
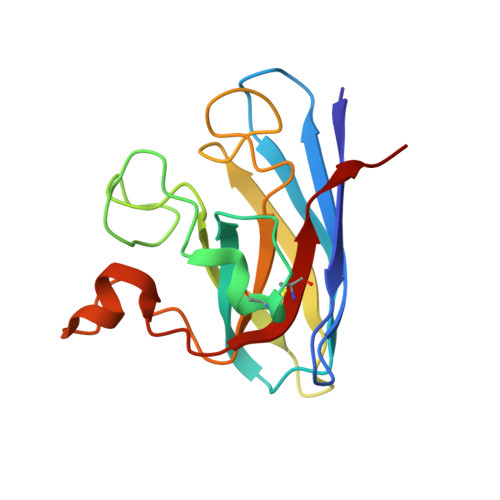
Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Chantadul, V., Wright, G.S.A., Amporndanai, K., Shahid, M., Antonyuk, S.V., Washbourn, G., Rogers, M., Roberts, N., Pye, M., O'Neill, P.M., Hasnain, S.S.(2020) Commun Biol 3: 97-97
- PubMed: 32139772
- DOI: https://doi.org/10.1038/s42003-020-0826-3
- Primary Citation of Related Structures:
6SPA, 6SPH, 6SPI, 6SPJ, 6SPK - PubMed Abstract:
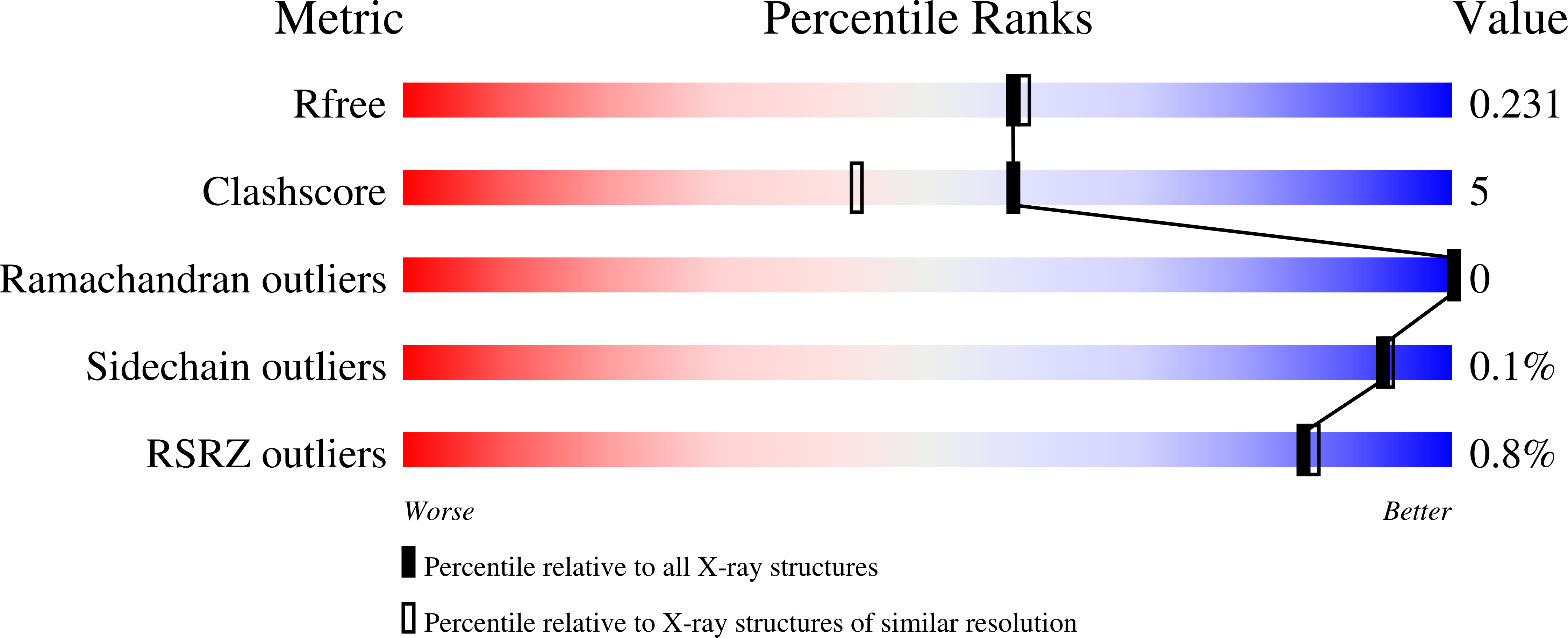
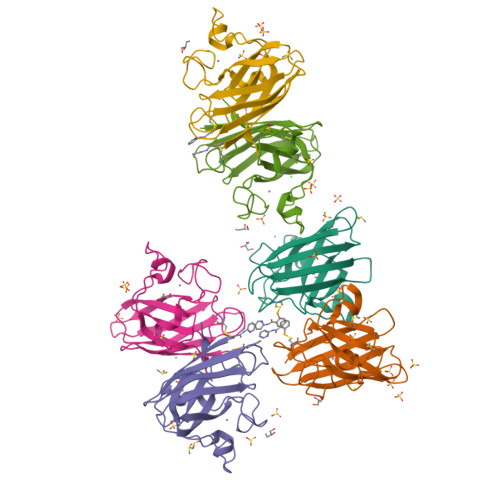
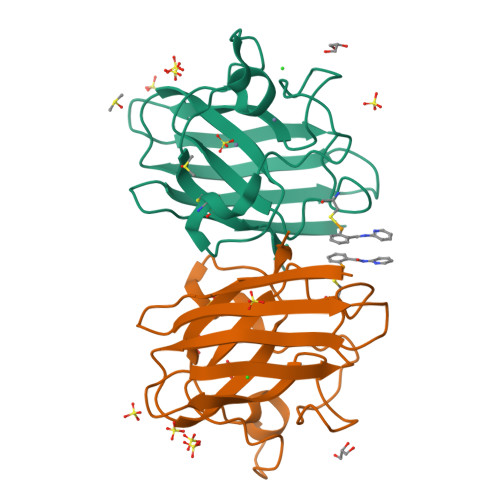
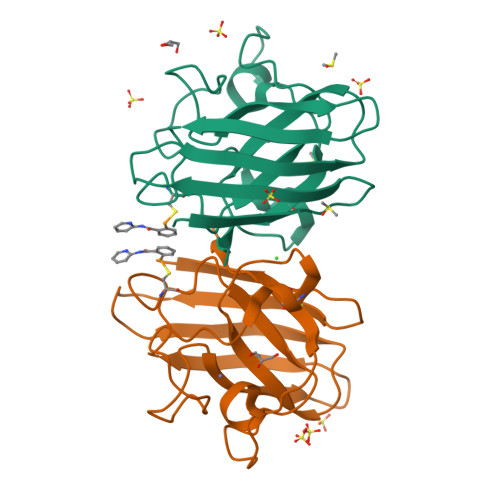
Mutations to the gene encoding superoxide dismutase-1 (SOD1) were the first genetic elements discovered that cause motor neuron disease (MND). These mutations result in compromised SOD1 dimer stability, with one of the severest and most common mutations Ala4Val (A4V) displaying a propensity to monomerise and aggregate leading to neuronal death. We show that the clinically used ebselen and related analogues promote thermal stability of A4V SOD1 when binding to Cys111 only. We have developed a A4V SOD1 differential scanning fluorescence-based assay on a C6S mutation background that is effective in assessing suitability of compounds. Crystallographic data show that the selenium atom of these compounds binds covalently to A4V SOD1 at Cys111 at the dimer interface, resulting in stabilisation. This together with chemical amenability for hit expansion of ebselen and its on-target SOD1 pharmacological chaperone activity holds remarkable promise for structure-based therapeutics for MND using ebselen as a template.
Organizational Affiliation:
Faculty of Health and Life Sciences, Molecular Biophysics Group, Institute of Integrative Biology, University of Liverpool, Liverpool, L69 7ZB, UK.