Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
Mainolfi, N., Ehara, T., Karki, R.G., Anderson, K., Mac Sweeney, A., Liao, S.M., Argikar, U.A., Jendza, K., Zhang, C., Powers, J., Klosowski, D.W., Crowley, M., Kawanami, T., Ding, J., April, M., Forster, C., Serrano-Wu, M., Capparelli, M., Ramqaj, R., Solovay, C., Cumin, F., Smith, T.M., Ferrara, L., Lee, W., Long, D., Prentiss, M., De Erkenez, A., Yang, L., Liu, F., Sellner, H., Sirockin, F., Valeur, E., Erbel, P., Ostermeier, D., Ramage, P., Gerhartz, B., Schubart, A., Flohr, S., Gradoux, N., Feifel, R., Vogg, B., Wiesmann, C., Maibaum, J., Eder, J., Sedrani, R., Harrison, R.A., Mogi, M., Jaffee, B.D., Adams, C.M.(2020) J Med Chem 63: 5697-5722
- PubMed: 32073845
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01870
- Primary Citation of Related Structures:
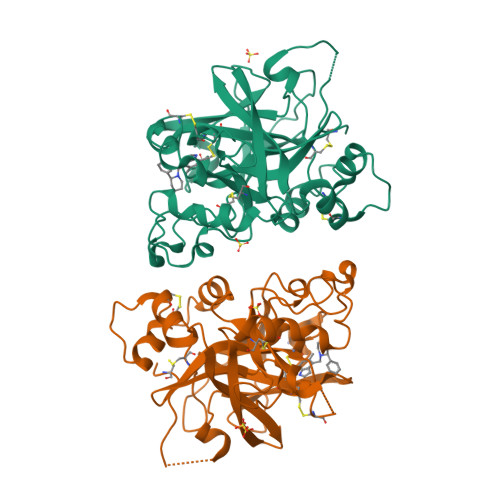
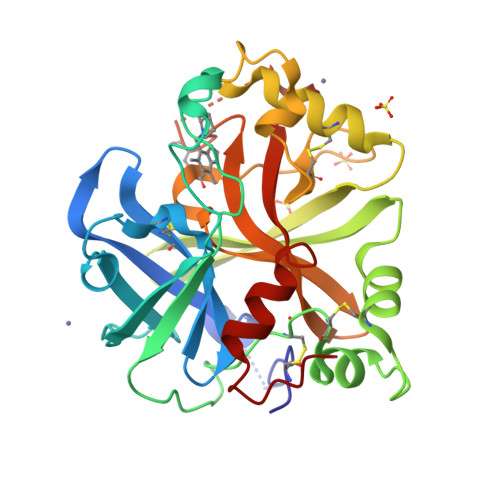
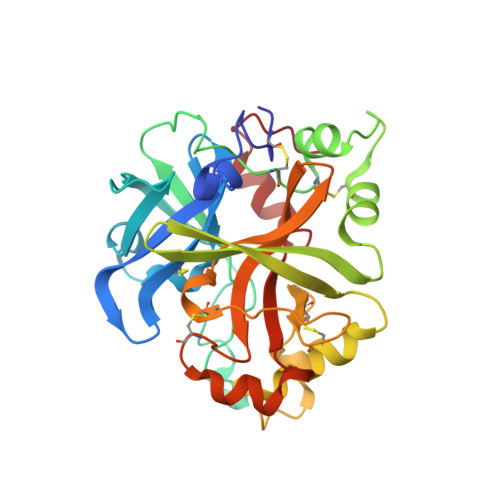
6T8U, 6T8V, 6T8W - PubMed Abstract:
The alternative pathway (AP) of the complement system is a key contributor to the pathogenesis of several human diseases including age-related macular degeneration, paroxysmal nocturnal hemoglobinuria (PNH), atypical hemolytic uremic syndrome (aHUS), and various glomerular diseases. The serine protease factor B (FB) is a key node in the AP and is integral to the formation of C3 and C5 convertase. Despite the prominent role of FB in the AP, selective orally bioavailable inhibitors, beyond our own efforts, have not been reported previously. Herein we describe in more detail our efforts to identify FB inhibitors by high-throughput screening (HTS) and leveraging insights from several X-ray cocrystal structures during optimization efforts. This work culminated in the discovery of LNP023 ( 41 ), which is currently being evaluated clinically in several diverse AP mediated indications.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States.