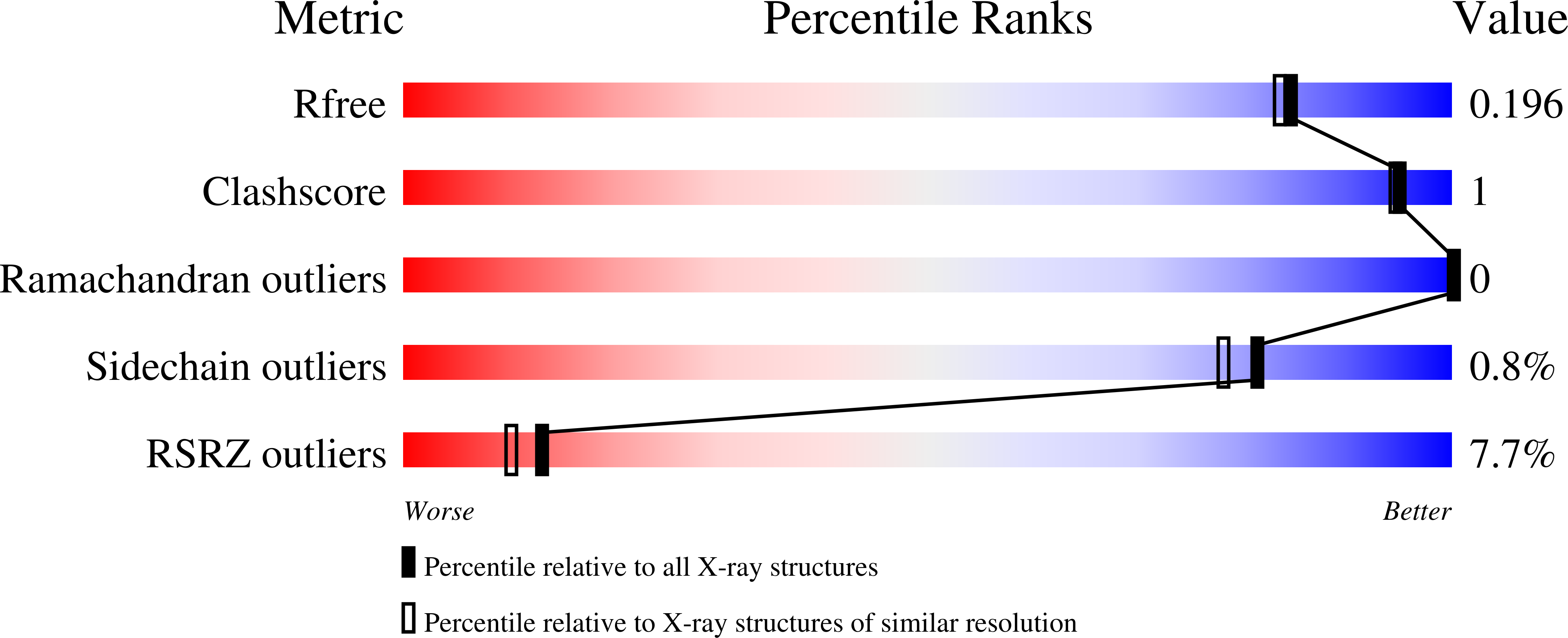
Substrate Promiscuity of a Paralytic Shellfish Toxin Amidinotransferase.
Lukowski, A.L., Mallik, L., Hinze, M.E., Carlson, B.M., Ellinwood, D.C., Pyser, J.B., Koutmos, M., Narayan, A.R.H.(2020) ACS Chem Biol 15: 626-631
- PubMed: 32058687
- DOI: https://doi.org/10.1021/acschembio.9b00964
- Primary Citation of Related Structures:
6U1R - PubMed Abstract:
Secondary metabolites are assembled by enzymes that often perform reactions with high selectivity and specificity. Many of these enzymes also tolerate variations in substrate structure, exhibiting promiscuity that enables various applications of a given biocatalyst. However, initial enzyme characterization studies frequently do not explore beyond the native substrates. This limited assessment of substrate scope contributes to the difficulty of identifying appropriate enzymes for specific synthetic applications. Here, we report the natural function of cyanobacterial SxtG, an amidinotransferase involved in the biosynthesis of paralytic shellfish toxins, and demonstrate its ability to modify a breadth of non-native substrates. In addition, we report the first X-ray crystal structure of SxtG, which provides rationale for this enzyme's substrate scope. Taken together, these data confirm the function of SxtG and exemplify its potential utility in biocatalytic synthesis.