Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Muslinkina, L., Gavrikov, A.S., Bozhanova, N.G., Mishin, A.S., Baranov, M.S., Meiler, J., Pletneva, N.V., Pletnev, V.Z., Pletnev, S.(2020) ACS Chem Biol 15: 2456-2465
- PubMed: 32809793
- DOI: https://doi.org/10.1021/acschembio.0c00440
- Primary Citation of Related Structures:
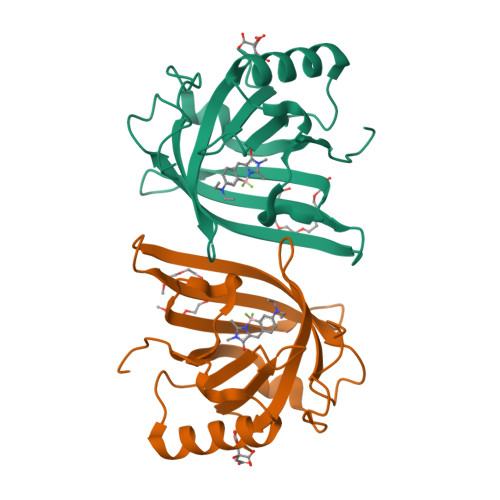
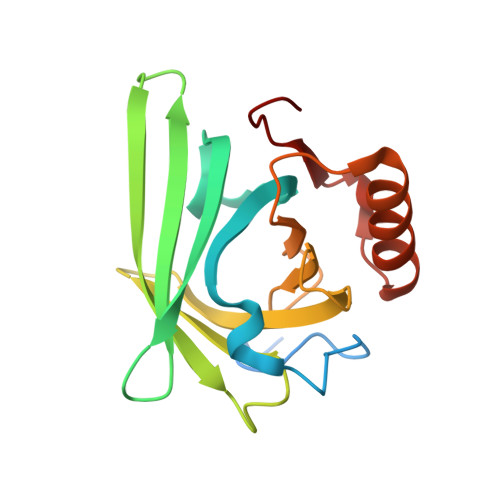
6UBO - PubMed Abstract:
Super-resolution fluorescent imaging in living cells remains technically challenging, largely due to the photodecomposition of fluorescent tags. The recently suggested protein-PAINT is the only super-resolution technique available for prolonged imaging of proteins in living cells. It is realized with complexes of fluorogen-activating proteins, expressed as fusions, and solvatochromic synthetic dyes. Once photobleached, the dye in the complex is replaced with a fresh fluorogen available in the sample. With suitable kinetics, this replacement creates fluorescence blinking required for attaining super-resolution and overcomes photobleaching associated with the loss of an irreplaceable fluorophore. Here we report on the rational design of two protein-PAINT tags based on the 1.58 Å crystal structure of the DiB1:M739 complex, an improved green-emitting DiB3/F74V:M739 and a new orange-emitting DiB3/F53L:M739. They outperform previously reported DiB-based tags to become best in class biomarkers for protein-PAINT. The new tags advance protein-PAINT from the proof-of-concept to a reliable tool suitable for prolonged super-resolution imaging of intracellular proteins in fixed and living cells and two-color PAINT-like nanoscopy with a single fluorogen.
Organizational Affiliation:
Basic Research Program, Frederick National Laboratory for Cancer Research, Argonne, Illinois 60439, United States.