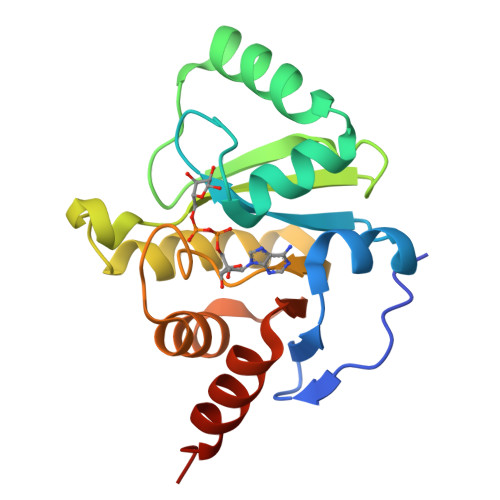
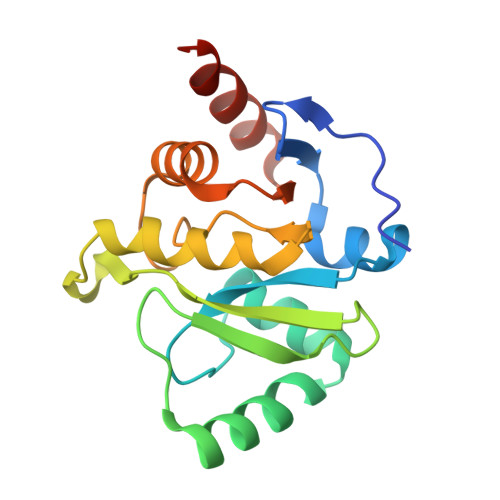
The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
Alhammad, Y.M.O., Kashipathy, M.M., Roy, A., Gagne, J.P., McDonald, P., Gao, P., Nonfoux, L., Battaile, K.P., Johnson, D.K., Holmstrom, E.D., Poirier, G.G., Lovell, S., Fehr, A.R.(2021) J Virol 95
- PubMed: 33158944
- DOI: https://doi.org/10.1128/JVI.01969-20
- Primary Citation of Related Structures:
6WOJ - PubMed Abstract:
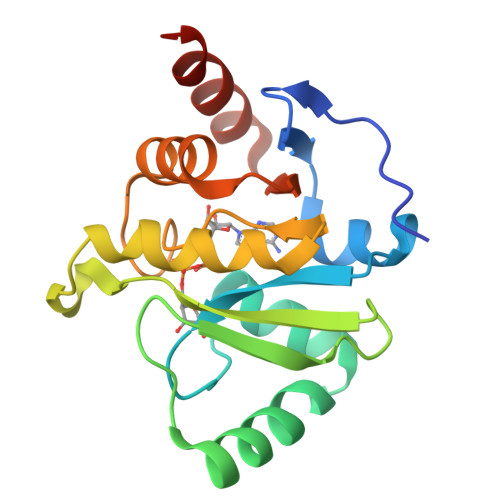
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other SARS-related CoVs encode 3 tandem macrodomains within nonstructural protein 3 (nsp3). The first macrodomain, Mac1, is conserved throughout CoVs and binds to and hydrolyzes mono-ADP-ribose (MAR) from target proteins. Mac1 likely counters host-mediated antiviral ADP-ribosylation, a posttranslational modification that is part of the host response to viral infections. Mac1 is essential for pathogenesis in multiple animal models of CoV infection, implicating it as a virulence factor and potential therapeutic target. Here, we report the crystal structure of SARS-CoV-2 Mac1 in complex with ADP-ribose. SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) Mac1 domains exhibit similar structural folds, and all 3 proteins bound to ADP-ribose with affinities in the low micromolar range. Importantly, using ADP-ribose-detecting binding reagents in both a gel-based assay and novel enzyme-linked immunosorbent assays (ELISAs), we demonstrated de-MARylating activity for all 3 CoV Mac1 proteins, with the SARS-CoV-2 Mac1 protein leading to a more rapid loss of substrate than the others. In addition, none of these enzymes could hydrolyze poly-ADP-ribose. We conclude that the SARS-CoV-2 and other CoV Mac1 proteins are MAR-hydrolases with similar functions, indicating that compounds targeting CoV Mac1 proteins may have broad anti-CoV activity. IMPORTANCE SARS-CoV-2 has recently emerged into the human population and has led to a worldwide pandemic of COVID-19 that has caused more than 1.2 million deaths worldwide. With no currently approved treatments, novel therapeutic strategies are desperately needed. All coronaviruses encode a highly conserved macrodomain (Mac1) that binds to and removes ADP-ribose adducts from proteins in a dynamic posttranslational process that is increasingly being recognized as an important factor that regulates viral infection. The macrodomain is essential for CoV pathogenesis and may be a novel therapeutic target. Thus, understanding its biochemistry and enzyme activity are critical first steps for these efforts. Here, we report the crystal structure of SARS-CoV-2 Mac1 in complex with ADP-ribose and describe its ADP-ribose binding and hydrolysis activities in direct comparison to those of SARS-CoV and MERS-CoV Mac1 proteins. These results are an important first step for the design and testing of potential therapies targeting this unique protein domain.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, Lawrence, Kansas, USA.