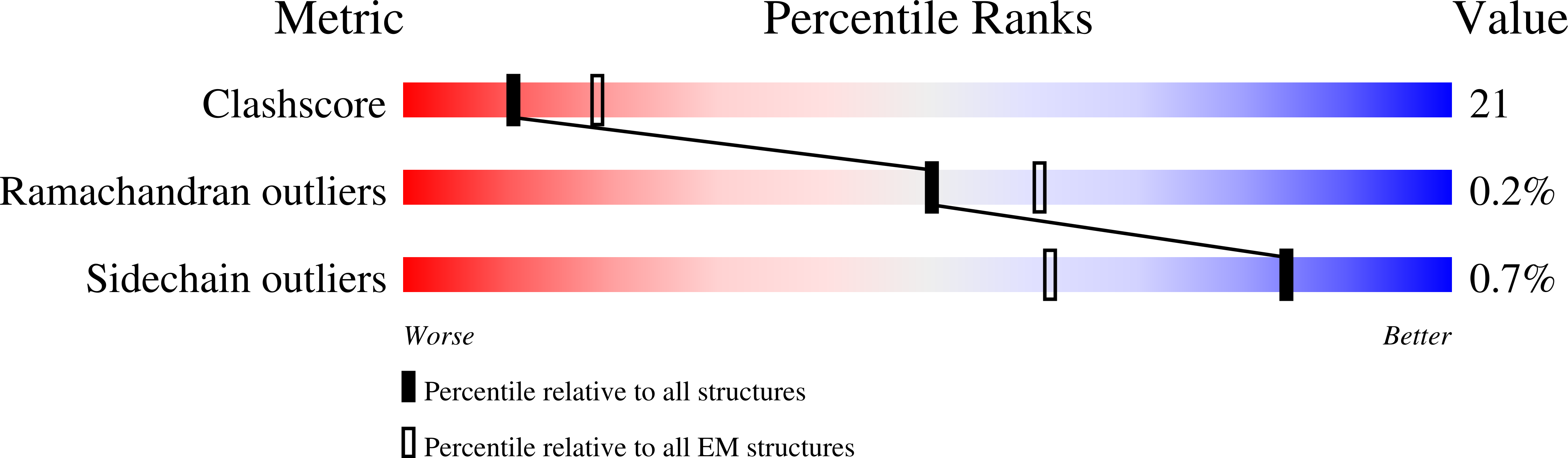Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
Li, S., Hsieh, K.Y., Su, S.C., Pintilie, G.D., Zhang, K., Chang, C.I.(2021) J Biol Chem 297: 101239-101239
- PubMed: 34563541
- DOI: https://doi.org/10.1016/j.jbc.2021.101239
- Primary Citation of Related Structures:
6WQH - PubMed Abstract:
The Lon AAA+ (adenosine triphosphatases associated with diverse cellular activities) protease (LonA) converts ATP-fuelled conformational changes into sufficient mechanical force to drive translocation of a substrate into a hexameric proteolytic chamber. To understand the structural basis for the substrate translocation process, we determined the cryo-electron microscopy (cryo-EM) structure of Meiothermus taiwanensis LonA (MtaLonA) in a substrate-engaged state at 3.6 Å resolution. Our data indicate that substrate interactions are mediated by the dual pore loops of the ATPase domains, organized in spiral staircase arrangement from four consecutive protomers in different ATP-binding and hydrolysis states. However, a closed AAA+ ring is maintained by two disengaged ADP-bound protomers transiting between the lowest and highest position. This structure reveals a processive rotary translocation mechanism mediated by LonA-specific nucleotide-dependent allosteric coordination among the ATPase domains, which is induced by substrate binding.
Organizational Affiliation:
MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale and Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.