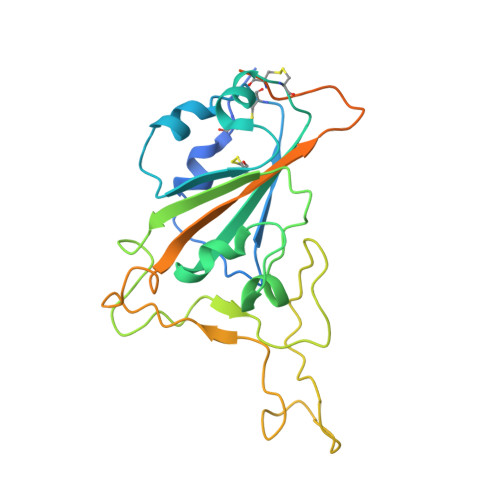
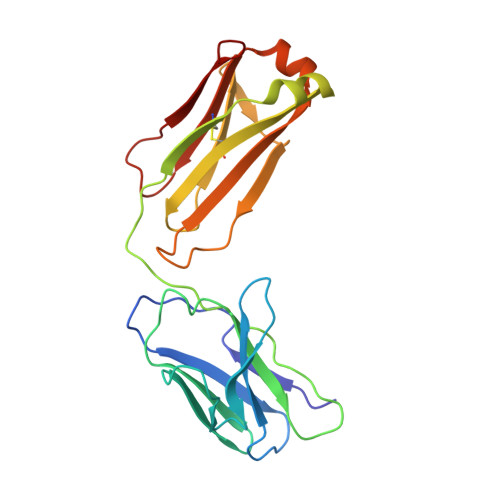
Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Hansen, J., Baum, A., Pascal, K.E., Russo, V., Giordano, S., Wloga, E., Fulton, B.O., Yan, Y., Koon, K., Patel, K., Chung, K.M., Hermann, A., Ullman, E., Cruz, J., Rafique, A., Huang, T., Fairhurst, J., Libertiny, C., Malbec, M., Lee, W.Y., Welsh, R., Farr, G., Pennington, S., Deshpande, D., Cheng, J., Watty, A., Bouffard, P., Babb, R., Levenkova, N., Chen, C., Zhang, B., Romero Hernandez, A., Saotome, K., Zhou, Y., Franklin, M., Sivapalasingam, S., Lye, D.C., Weston, S., Logue, J., Haupt, R., Frieman, M., Chen, G., Olson, W., Murphy, A.J., Stahl, N., Yancopoulos, G.D., Kyratsous, C.A.(2020) Science 369: 1010-1014
- PubMed: 32540901
- DOI: https://doi.org/10.1126/science.abd0827
- Primary Citation of Related Structures:
6XDG - PubMed Abstract:
Neutralizing antibodies have become an important tool in treating infectious diseases. Recently, two separate approaches yielded successful antibody treatments for Ebola-one from genetically humanized mice and the other from a human survivor. Here, we describe parallel efforts using both humanized mice and convalescent patients to generate antibodies against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein, which yielded a large collection of fully human antibodies that were characterized for binding, neutralization, and three-dimensional structure. On the basis of these criteria, we selected pairs of highly potent individual antibodies that simultaneously bind the receptor binding domain of the spike protein, thereby providing ideal partners for a therapeutic antibody cocktail that aims to decrease the potential for virus escape mutants that might arise in response to selective pressure from a single-antibody treatment.
Organizational Affiliation:
Regeneron Pharmaceuticals, Inc., Tarrytown, NY 10591, USA.